 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSMUA_Achr1P06280_001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Zingiberales; Musaceae; Musa
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 413aa MW: 46077.3 Da PI: 9.8332 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 101.8 | 4.3e-32 | 237 | 290 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
pr+rWt++LH+rFv+ave LGG+e+AtPk++lelm+vk+Ltl+hvkSHLQ+YR+
GSMUA_Achr1P06280_001 237 PRMRWTTTLHARFVHAVELLGGHERATPKSVLELMDVKDLTLAHVKSHLQMYRT 290
9****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.79E-15 | 234 | 291 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 1.4E-28 | 235 | 291 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 2.8E-23 | 237 | 290 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 2.8E-7 | 238 | 289 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009944 | Biological Process | polarity specification of adaxial/abaxial axis | ||||
| GO:0048481 | Biological Process | plant ovule development | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 413 aa Download sequence Send to blast |
MAIRISKFQV YPQIHARRRR SSHKLKSFVP KEVICCCCCK KKKKKKRSEV LEQKLMELFP 60 AQPDLSLQIS PPNTTQASAW RKADENMDLG FWSSSSSYNN RKSITGPCTA MAGETAFGLS 120 LASPGATDCS SARDSLLLRP HPHYQHHHLH FHHHHHPLLQ EAGFREDLGV LKPIRGIPVY 180 QHKPSFPLVP LHQQQQQQQP CDSSSTSNFS PFAATQGLSR SRFLPTRFPG KRSMRAPRMR 240 WTTTLHARFV HAVELLGGHE RATPKSVLEL MDVKDLTLAH VKSHLQMYRT VKNTDKPAVS 300 SGQSDGFENG STGEICDDNS PDNPDQRGPG STAAKMMQHG VGLWSNSSRG GCFTGTPGES 360 TAGSMHFCFL FSFLQKDLRS KSLEMYSDMN SSCLSETLSS SMPNLEFTLG RSQ |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 2e-16 | 238 | 292 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 2e-16 | 238 | 292 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_A | 2e-16 | 238 | 292 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 2e-16 | 238 | 292 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 2e-16 | 238 | 292 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 2e-16 | 238 | 292 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 2e-16 | 238 | 292 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 2e-16 | 238 | 292 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 2e-16 | 238 | 292 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 2e-16 | 238 | 292 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 2e-16 | 238 | 292 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 2e-16 | 238 | 292 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 40 | 46 | KKKKKKR |
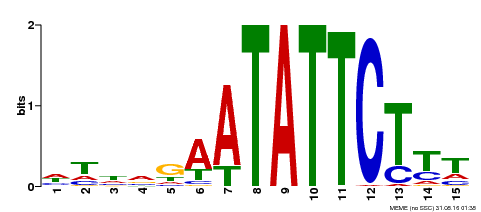
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00174 | DAP | Transfer from AT1G32240 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009395186.1 | 0.0 | PREDICTED: probable transcription factor KAN2 | ||||
| TrEMBL | M0RXI6 | 0.0 | M0RXI6_MUSAM; Uncharacterized protein | ||||
| STRING | GSMUA_Achr1P06280_001 | 0.0 | (Musa acuminata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP4721 | 36 | 65 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G32240.1 | 3e-57 | G2-like family protein | ||||



