 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSMUA_Achr1P07110_001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Zingiberales; Musaceae; Musa
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 486aa MW: 52446.1 Da PI: 9.7159 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 16.5 | 2.4e-05 | 112 | 134 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
+ C+ C+k F r nL+ H r H
GSMUA_Achr1P07110_001 112 FICEVCNKGFQREQNLQLHRRGH 134
89*******************88 PP
| |||||||
| 2 | zf-C2H2 | 11.9 | 0.00069 | 188 | 210 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
+kC +C+k++ +s+ k H + +
GSMUA_Achr1P07110_001 188 WKCDKCSKRYAVQSDWKAHSKIC 210
58*****************9876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF57667 | 2.18E-7 | 111 | 134 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 5.2E-6 | 111 | 134 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 10.99 | 112 | 134 | IPR007087 | Zinc finger, C2H2 |
| Pfam | PF12171 | 2.3E-5 | 112 | 134 | IPR022755 | Zinc finger, double-stranded RNA binding |
| SMART | SM00355 | 0.0052 | 112 | 134 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 114 | 134 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 3.1E-4 | 176 | 209 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 2.18E-7 | 183 | 208 | No hit | No description |
| SMART | SM00355 | 140 | 188 | 208 | IPR015880 | Zinc finger, C2H2-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 486 aa Download sequence Send to blast |
MKAASSSTHF FGIREDGQHS HQQPQQPPQP PPPQELPATA SSANTPHSAA PPKKRRNLPG 60 NPSKYLSLFS FTSFLCSLSL NLGLYMRSIM ITDPDAEVIA LSPKTLMATN RFICEVCNKG 120 FQREQNLQLH RRGHNLPWKL KQKNPNEVRR RVYLCPEPTC VHHDPSRALG DLTGIKKHFC 180 RKHGEKKWKC DKCSKRYAVQ SDWKAHSKIC GTREYRCDCG TLFSRRDSFI THRAFCDALA 240 QESARLPGGI NTIGSHLYGG RGMNLGLPQL SSLQDQAQPS ADLLQLRGSS GTGHNAGGGT 300 EAMSLFAGDL MSNHTDTTMS SLYNPSVHAE SVVPQMSATA LLQKAAQMGA TSSGGSGSSM 360 LRGLTASFSG GGGGEISRTG VGNENHFQDI MNSLANGNGG VFGGFNQGLD GMGENKLHRN 420 LSMGGFGGSD RLTRDFLGVG GMMRSMVGGI PQREQHHLGI DTTSSMDHSE MKSGSSTRSF 480 GGGRLQ |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5b3h_C | 2e-34 | 184 | 246 | 3 | 65 | Zinc finger protein JACKDAW |
| 5b3h_F | 2e-34 | 184 | 246 | 3 | 65 | Zinc finger protein JACKDAW |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000305}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00142 | DAP | Transfer from AT1G14580 | Download |
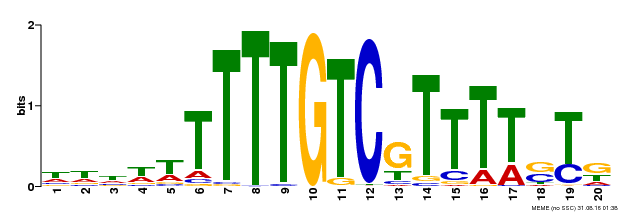
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK374308 | 1e-175 | AK374308.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv3060C18. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009394230.1 | 1e-177 | PREDICTED: protein indeterminate-domain 5, chloroplastic-like | ||||
| Refseq | XP_009394238.1 | 1e-177 | PREDICTED: protein indeterminate-domain 5, chloroplastic-like | ||||
| Refseq | XP_009394246.1 | 1e-177 | PREDICTED: protein indeterminate-domain 5, chloroplastic-like | ||||
| Swissprot | Q8RWX7 | 1e-110 | IDD6_ARATH; Protein indeterminate-domain 6, chloroplastic | ||||
| TrEMBL | M0RXR9 | 0.0 | M0RXR9_MUSAM; Uncharacterized protein | ||||
| STRING | GSMUA_Achr1P07110_001 | 0.0 | (Musa acuminata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP202 | 38 | 320 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G02080.1 | 1e-109 | indeterminate(ID)-domain 4 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



