 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSMUA_Achr2P01820_001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Zingiberales; Musaceae; Musa
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 1018aa MW: 113101 Da PI: 6.4704 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 130.1 | 8.2e-41 | 124 | 200 | 1 | 77 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S-- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkq 77
+CqvegC+adls+ k+yhrrhkvCe+h+ka++++v++ +qrfCqqCsrfh l+efDe+krsCrrrLa+hn+rrrk+q
GSMUA_Achr2P01820_001 124 ACQVEGCDADLSDSKDYHRRHKVCEMHAKASTAVVRNASQRFCQQCSRFHLLEEFDEGKRSCRRRLAGHNRRRRKTQ 200
5**************************************************************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 3.1E-34 | 118 | 186 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.556 | 122 | 199 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 5.49E-38 | 123 | 204 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 2.6E-30 | 125 | 198 | IPR004333 | Transcription factor, SBP-box |
| CDD | cd00204 | 1.62E-9 | 717 | 848 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 2.5E-6 | 742 | 845 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 5.51E-8 | 743 | 851 | IPR020683 | Ankyrin repeat-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1018 aa Download sequence Send to blast |
MEARIGGENH QFVAAGTSKL NRIGKKSLEW DLNDWRWDGD RFVASPLNSI PEDCRNKQLH 60 QDTAITLVSN CSSSSSEGVD YRLVDKEKGE AEKRRRIVVG EEDESFGKTG KKSILLGGNS 120 SHPACQVEGC DADLSDSKDY HRRHKVCEMH AKASTAVVRN ASQRFCQQCS RFHLLEEFDE 180 GKRSCRRRLA GHNRRRRKTQ LDVNVNGNSL IDEQACGYLL ISLLRILSNL QSANSDQSQD 240 QELLTNFLGN LATFANSFDP SGLSRLLQAS QDPQKLVTSS GISTDVVITS VPNGVREQES 300 GNPLCSTAVM TCITGTQDPL RQTDHVTSVS VTTVDVPSKE RVASPVIMDF DLNNAYSDTR 360 DCEEGRMNPA TLLSTRMDSS NCPSWLLQSS HQSSPPQTSG NSDTCNQSQT SSHGGAQCRT 420 DRIIFKLFGK DPNDLPLALR TQILDWLSSG PTDIESYIRP GCIVLTIYLQ QAESAWVQLS 480 HDLSSNLSRL LHDSNDFWTT GWIFARVQNC AVFINDGQVV LDMPFPIGDF NHCQGLSVTP 540 IAVACSTKVK FTVKGFNLVQ PTSGLLCSFD GKYLVQETTQ ALVEGTGRDA GHDLSQCLSF 600 TCLLPDVTGR GFIEFEDCGL CNGFFPFIVA EEDVCSEIRM LENAINIASC DGQLQERTDA 660 ANARNQALDF INELGWLLRK NHMRSASEGT KFSQNTFPLR RFRHLMSFAM SREWSAVVKK 720 LLDILFSGTV DADRQSPTEL ALSENLLHSA VQMNSRPMVE LLLRYAPVKA SKETDVDRFL 780 FRPDMLGPLG ITPLHVAASS NGAESILDAL TDDPELLGIK AWNNVRDCIG FTPEDYALAQ 840 GHDSYIRLVQ KKIDKQHHQS QVVLNIPGVV SYELVDALKS GKPNLFQITK SCLSRERQPY 900 CNRCSQKIAY PNSVARTILY RPVMLSLVGI AAVCVCMGLL FKTPPQVFYV FPSFRWELLD 960 YGFISSFLVV DRMAQHQGKL VREILVEFFL LFIELNASSK HVSHCSGKQR DAGNIMMA |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 7e-30 | 118 | 198 | 4 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 92 | 96 | KRRRI |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00633 | PBM | Transfer from PK06791.1 | Download |
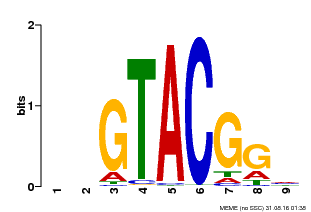
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009381220.1 | 0.0 | PREDICTED: squamosa promoter-binding-like protein 6 isoform X1 | ||||
| TrEMBL | M0S4C3 | 0.0 | M0S4C3_MUSAM; Uncharacterized protein | ||||
| STRING | GSMUA_Achr2P01820_001 | 0.0 | (Musa acuminata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1998 | 38 | 92 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G47070.1 | 0.0 | squamosa promoter binding protein-like 1 | ||||



