 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSMUA_Achr2P04340_001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Zingiberales; Musaceae; Musa
|
||||||||
| Family | MIKC_MADS | ||||||||
| Protein Properties | Length: 213aa MW: 24504.1 Da PI: 9.1287 | ||||||||
| Description | MIKC_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 96 | 1.6e-30 | 10 | 59 | 2 | 51 |
---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TTSEEEEEE- CS
SRF-TF 2 rienksnrqvtfskRrngilKKAeELSvLCdaevaviifsstgklyeyss 51
rien rqvtfskRrng+lKKA+ELSvLCdaevavi+fs++gklye+ss
GSMUA_Achr2P04340_001 10 RIENAASRQVTFSKRRNGLLKKAYELSVLCDAEVAVIVFSPRGKLYEFSS 59
8***********************************************96 PP
| |||||||
| 2 | K-box | 75.4 | 1.5e-25 | 83 | 170 | 10 | 97 |
K-box 10 eeakaeslqqelakLkkeienLqreqRhllGedLesLslkeLqqLeqqLekslkkiRskKnellleqieelqkkekelqeenkaLrkk 97
++++++++ e++ + k+ie ++ ++R+llG++Les+slkeL +Le q+e+sl kiR +K+ +l+eqi +l+ k + l een +L+++
GSMUA_Achr2P04340_001 83 TDSDMQQWISEATDIAKKIEVIEAQKRKLLGDGLESCSLKELCELEAQMERSLHKIRGRKQYVLTEQIAQLKDKVRVLLEENVKLHRE 170
68899********************************************************************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00432 | 1.7E-41 | 1 | 60 | IPR002100 | Transcription factor, MADS-box |
| PROSITE profile | PS50066 | 31.707 | 1 | 61 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 1.7E-30 | 3 | 23 | IPR002100 | Transcription factor, MADS-box |
| PROSITE pattern | PS00350 | 0 | 3 | 57 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00265 | 1.52E-41 | 3 | 75 | No hit | No description |
| SuperFamily | SSF55455 | 3.27E-32 | 3 | 74 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 5.5E-28 | 10 | 57 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 1.7E-30 | 23 | 38 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 1.7E-30 | 38 | 59 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF01486 | 5.1E-25 | 85 | 170 | IPR002487 | Transcription factor, K-box |
| PROSITE profile | PS51297 | 13.356 | 87 | 183 | IPR002487 | Transcription factor, K-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009838 | Biological Process | abscission | ||||
| GO:0009909 | Biological Process | regulation of flower development | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0080187 | Biological Process | floral organ senescence | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 213 aa Download sequence Send to blast |
MVRGKTEMRR IENAASRQVT FSKRRNGLLK KAYELSVLCD AEVAVIVFSP RGKLYEFSSS 60 SVERTIDRYK MHTKDANIMD KGTDSDMQQW ISEATDIAKK IEVIEAQKRK LLGDGLESCS 120 LKELCELEAQ MERSLHKIRG RKQYVLTEQI AQLKDKVRVL LEENVKLHRE RHIEPQLQLA 180 STRDVGPKNH GNQEITEVET ELVIGRPGIR EDK |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 3mu6_A | 7e-18 | 3 | 72 | 2 | 71 | Myocyte-specific enhancer factor 2A |
| 3mu6_B | 7e-18 | 3 | 72 | 2 | 71 | Myocyte-specific enhancer factor 2A |
| 3mu6_C | 7e-18 | 3 | 72 | 2 | 71 | Myocyte-specific enhancer factor 2A |
| 3mu6_D | 7e-18 | 3 | 72 | 2 | 71 | Myocyte-specific enhancer factor 2A |
| 5f28_A | 8e-18 | 1 | 69 | 1 | 69 | MEF2C |
| 5f28_B | 8e-18 | 1 | 69 | 1 | 69 | MEF2C |
| 5f28_C | 8e-18 | 1 | 69 | 1 | 69 | MEF2C |
| 5f28_D | 8e-18 | 1 | 69 | 1 | 69 | MEF2C |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor active in flowering time control. May control internode elongation and promote floral transition phase. May act upstream of the floral regulators MADS1, MADS14, MADS15 and MADS18 in the floral induction pathway. {ECO:0000269|PubMed:15144377, ECO:0000269|PubMed:17166135}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00576 | DAP | Transfer from AT5G62165 | Download |
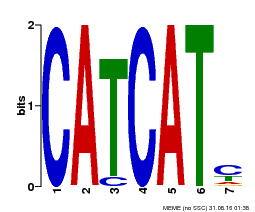
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | EU244589 | 2e-98 | EU244589.1 Musa acuminata cultivar Mas SOC1-like protein gene, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009384275.1 | 1e-156 | PREDICTED: MADS-box protein AGL42-like isoform X1 | ||||
| Refseq | XP_018678105.1 | 1e-156 | PREDICTED: MADS-box protein AGL42-like isoform X1 | ||||
| Swissprot | Q9XJ60 | 3e-69 | MAD50_ORYSJ; MADS-box transcription factor 50 | ||||
| TrEMBL | M0S525 | 1e-154 | M0S525_MUSAM; Uncharacterized protein | ||||
| STRING | GSMUA_Achr2P04340_001 | 1e-155 | (Musa acuminata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1413 | 33 | 79 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G62165.3 | 7e-64 | AGAMOUS-like 42 | ||||



