 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSMUA_Achr2P14750_001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Zingiberales; Musaceae; Musa
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 286aa MW: 32310.2 Da PI: 4.3446 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 60 | 3.7e-19 | 81 | 134 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k+++++ eq+++Le+ Fe +++++ e++ +LA+ lgL+ rqV vWFqNrRa++k
GSMUA_Achr2P14750_001 81 KKRRLSVEQVKALEKNFEVENKLDPERKLRLAQDLGLQPRQVAVWFQNRRARWK 134
456899***********************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 128.6 | 2.6e-41 | 80 | 172 | 1 | 93 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLr 88
ekkrrls eqvk+LE++Fe e+kL+perK +la++LglqprqvavWFqnrRAR+ktkqlE+dy aLk+++dalk + ++L++++e+L
GSMUA_Achr2P14750_001 80 EKKRRLSVEQVKALEKNFEVENKLDPERKLRLAQDLGLQPRQVAVWFQNRRARWKTKQLERDYGALKARHDALKLDCDALSRDNEALL 167
69*************************************************************************************9 PP
HD-ZIP_I/II 89 eelke 93
+e++e
GSMUA_Achr2P14750_001 168 AEIAE 172
99876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.24E-19 | 75 | 138 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.135 | 76 | 136 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.0E-18 | 79 | 140 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 1.7E-16 | 81 | 134 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 7.83E-18 | 81 | 137 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 2.6E-20 | 83 | 143 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 3.0E-5 | 107 | 116 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 111 | 134 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 3.0E-5 | 116 | 132 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 2.0E-17 | 136 | 177 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009637 | Biological Process | response to blue light | ||||
| GO:0030308 | Biological Process | negative regulation of cell growth | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0048510 | Biological Process | regulation of timing of transition from vegetative to reproductive phase | ||||
| GO:0048573 | Biological Process | photoperiodism, flowering | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 286 aa Download sequence Send to blast |
MVYKENLALS PSPYQVTLSL SLSLSLVDYL VFSLLLQSDL FYLFSVEEKG KYEAVLDGTE 60 ELDGCEEEEP CGTGLSGLGE KKRRLSVEQV KALEKNFEVE NKLDPERKLR LAQDLGLQPR 120 QVAVWFQNRR ARWKTKQLER DYGALKARHD ALKLDCDALS RDNEALLAEI AELKAKLADS 180 GMDAMESETK AVEEERPPLI HKDGASDSDS SVVFNDETSP HAGTVLHQHF LMGFRSQCPF 240 LPFEDKAQAE KGYLDEELLA GEELCSSLFS EQQAPNFSWY CSDEWD |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 128 | 136 | RRARWKTKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may function as a negative regulator of the flowering time response to photoperiod. May act to repress cell expansion during plant development. {ECO:0000269|PubMed:14623244}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00051 | PBM | Transfer from AT4G40060 | Download |
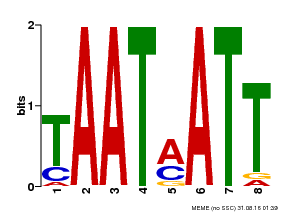
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009388778.1 | 1e-177 | PREDICTED: homeobox-leucine zipper protein ATHB-16 | ||||
| Swissprot | Q940J1 | 4e-58 | ATB16_ARATH; Homeobox-leucine zipper protein ATHB-16 | ||||
| TrEMBL | M0S816 | 0.0 | M0S816_MUSAM; Uncharacterized protein | ||||
| STRING | GSMUA_Achr2P14750_001 | 0.0 | (Musa acuminata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3009 | 34 | 82 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G40060.1 | 2e-50 | homeobox protein 16 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



