 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSMUA_Achr2P15140_001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Zingiberales; Musaceae; Musa
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 739aa MW: 82083.3 Da PI: 6.573 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 59 | 7.5e-19 | 16 | 71 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ +++++ q++eLe+lF+ + +p++++r +L++ lgL+ rq+k+WFqNrR+++k
GSMUA_Achr2P15140_001 16 KKRYHRHNPRQIQELESLFKVCLHPDEKQRLQLSRDLGLEPRQIKFWFQNRRTQMK 71
688899***********************************************998 PP
| |||||||
| 2 | START | 173.3 | 1.5e-54 | 219 | 441 | 3 | 206 |
HHHHHHHHHHHHHHC-TT-EEEE......EXCCTTEEEEEEESSS......SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S.. CS
START 3 aeeaaqelvkkalaeepgWvkss......esengdevlqkfeeskv.....dsgealrasgvvdmvlallveellddkeqWdetla.. 77
a a++e++++++ +ep+Wvkss + e++d q++ + ++ea+r+s++v+m +++l ++d + +W e ++
GSMUA_Achr2P15140_001 219 ATGAMEEVIRLVQTDEPLWVKSSdgrdilQLETYDSMFQRLGR--QlkfpgTRIEASRDSALVIMGAMTLIDMFMDAS-KWAELFPti 303
66799********************977777777777777755..256999***************************.****99999 PP
..EEEEEEEECTT......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--.-TTSEE-EESSEEEEEEE CS
START 78 ..kaetlevissg......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppesssvvRaellpSgiliep 156
ka t+ev+ g g l lm+ e q+lsp+vp R+f f+Ry++q + g+wv++dvSvd ++++ + + R+++lpSg+lie+
GSMUA_Achr2P15140_001 304 vsKARTIEVLAAGmagsrsGSLLLMYEEIQVLSPVVPmREFCFLRYCQQIEPGVWVVADVSVDYPRDNR-LALSSRSRRLPSGYLIEE 390
99******************************************************************9.7***************** PP
ECTCEEEEEEEE-EE--SSXX.HHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 157 ksnghskvtwvehvdlkgrlp.hwllrslvksglaegaktwvatlqrqcek 206
++ng++k+twveh++ +++ p h l+r l++sg+ +ga++w+a+lqr ce+
GSMUA_Achr2P15140_001 391 MPNGYTKLTWVEHMEIEDKNPiHILFRDLINSGMLFGAQRWLAALQRMCER 441
*************************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 1.6E-19 | 3 | 71 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 1.67E-18 | 7 | 71 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 16.52 | 13 | 73 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 4.4E-16 | 14 | 77 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 3.78E-16 | 15 | 71 | No hit | No description |
| Pfam | PF00046 | 2.1E-16 | 16 | 71 | IPR001356 | Homeobox domain |
| PROSITE pattern | PS00027 | 0 | 48 | 71 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 51.336 | 208 | 444 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 1.75E-35 | 210 | 442 | No hit | No description |
| CDD | cd08875 | 1.08E-105 | 212 | 440 | No hit | No description |
| SMART | SM00234 | 2.3E-47 | 217 | 441 | IPR002913 | START domain |
| Pfam | PF01852 | 1.1E-46 | 221 | 441 | IPR002913 | START domain |
| Gene3D | G3DSA:3.30.530.20 | 6.6E-10 | 277 | 440 | IPR023393 | START-like domain |
| SuperFamily | SSF55961 | 2.91E-21 | 461 | 698 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009828 | Biological Process | plant-type cell wall loosening | ||||
| GO:0010091 | Biological Process | trichome branching | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0000976 | Molecular Function | transcription regulatory region sequence-specific DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 739 aa Download sequence Send to blast |
MDSGDEQDVP NSQSRKKRYH RHNPRQIQEL ESLFKVCLHP DEKQRLQLSR DLGLEPRQIK 60 FWFQNRRTQM KATAALFSSF QRADNCLLRA ENDKIRCENI AMREALKNVI CPSCGGPPPD 120 DDSYFDEQKL RMDNTRLKEE LDRVSNLASK YLGRPLTQLP PVQPVSLSSL DLSAGGYGDA 180 GISPSLDLDL LCRSSSSAFP FQCPSTVSDL EKPLMVEMAT GAMEEVIRLV QTDEPLWVKS 240 SDGRDILQLE TYDSMFQRLG RQLKFPGTRI EASRDSALVI MGAMTLIDMF MDASKWAELF 300 PTIVSKARTI EVLAAGMAGS RSGSLLLMYE EIQVLSPVVP MREFCFLRYC QQIEPGVWVV 360 ADVSVDYPRD NRLALSSRSR RLPSGYLIEE MPNGYTKLTW VEHMEIEDKN PIHILFRDLI 420 NSGMLFGAQR WLAALQRMCE RFACLNVAGL PARDIGVAPS PDGKRSMMKL AQRMLSSFCA 480 NVGASNGHQW NTISGLNDVG VRVTIHKTTD AGRPDGIVLN AATSMWLPIS SEKVFGFFKD 540 ERTRSQWDVL SNGNTLQEVA HITNGSHPGN CISLLRVCSF SLGLNSGQNT MLILQECCTD 600 AYGSVIVYSP VDLPAINIVM SGEDPSYIPI LPSGFTILPD GRAAAGASSS SNPMVGSSGS 660 LLTVAFQILM SGLPSAKLNL ESVMTVNNLI GTTVQQIKSA LNCTCINSCN FLKDLCSWLN 720 GWFLHISWLH LFVQMLPKY |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00232 | DAP | Transfer from AT1G73360 | Download |
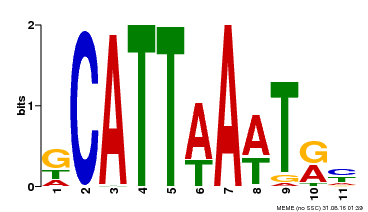
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009388746.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein ROC8-like | ||||
| Swissprot | Q69T58 | 0.0 | ROC8_ORYSJ; Homeobox-leucine zipper protein ROC8 | ||||
| TrEMBL | M0S855 | 0.0 | M0S855_MUSAM; Uncharacterized protein | ||||
| STRING | GSMUA_Achr2P15140_001 | 0.0 | (Musa acuminata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2912 | 37 | 70 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G73360.1 | 0.0 | homeodomain GLABROUS 11 | ||||



