 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSMUA_Achr2P18140_001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Zingiberales; Musaceae; Musa
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 464aa MW: 50614 Da PI: 6.7387 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 39.8 | 8e-13 | 308 | 353 | 4 | 54 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksL 54
+h e+Er+RR+++N++f Lr ++P+ K++Ka+ L Av YI++L
GSMUA_Achr2P18140_001 308 NHVEAERQRREKLNQRFYALRAVVPNV-----SKMDKASLLGDAVSYINEL 353
799***********************6.....5***************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 4.2E-42 | 63 | 217 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| PROSITE profile | PS50888 | 17.301 | 304 | 353 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 2.67E-15 | 307 | 358 | No hit | No description |
| SuperFamily | SSF47459 | 5.23E-18 | 307 | 369 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 2.7E-17 | 308 | 367 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 2.4E-10 | 308 | 353 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.4E-16 | 310 | 359 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006952 | Biological Process | defense response | ||||
| GO:0009718 | Biological Process | anthocyanin-containing compound biosynthetic process | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043425 | Molecular Function | bHLH transcription factor binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 464 aa Download sequence Send to blast |
MNLWADDNAS MMEAFMASVD LPGFSWAAVP PTPTPPCAAA SSSLDHPAAA ATPAPAYFSQ 60 ETLQQRLQAL IEGARESWTY GIFWQSSVDA ATGASFLGWG DGYYKGYEED KRKQRVAGAA 120 SAAEQEHRKC WFFLVSMTQS FVNGGGLPGQ AFYTGAPAWV TGADRLAAAP CDRARQAQLF 180 GLQTMVCVPV GSGVLELGST DVVFHSPEIM GKIRVLFNFS SPDAPSFQRT HNQPQQQLHQ 240 SSASDPHTHL FVSKKFNLSE FASNGSRNKR STGATSRVSN SDEGMISFSS ASARPPSDPA 300 NGREEPLNHV EAERQRREKL NQRFYALRAV VPNVSKMDKA SLLGDAVSYI NELRSKLETL 360 EIDKEEMMNG SGGDRCHGVE LEVKILGSEA MIRLQCLKRN HPAAKLMAAI RDLDLDVHYA 420 SVSVVEDLMI QQATVKMSPS RTYTPEQLRA ALYSKLAAEA PISR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4rqw_A | 7e-48 | 58 | 220 | 4 | 195 | Transcription factor MYC3 |
| 4rqw_B | 7e-48 | 58 | 220 | 4 | 195 | Transcription factor MYC3 |
| 4rs9_A | 7e-48 | 58 | 220 | 4 | 195 | Transcription factor MYC3 |
| 4yz6_A | 7e-48 | 58 | 220 | 4 | 195 | Transcription factor MYC3 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that binds to the G-box motif (5'-AACGTG-3') found in a number of promoters of jasmonate-induced genes (PubMed:15231736). Transcription activator involved in the transcriptional regulation of terpene biosynthesis in glandular trichomes (PubMed:24884371, PubMed:30518626). Binds to the promoter of the linalool synthase TPS5 and promotes TPS5 gene transactivation (PubMed:24884371). Acts synergistically with EOT1 in the transactivation of TPS5 (PubMed:24884371). Involved in type VI glandular trichome development (PubMed:30518626). Involved in the activation of terpene synthases required for volatile mono- and sesquiterpenes synthesis by the glandular cells of type VI trichomes (PubMed:30518626). {ECO:0000269|PubMed:15231736, ECO:0000269|PubMed:24884371, ECO:0000269|PubMed:30518626}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00086 | PBM | Transfer from AT4G17880 | Download |
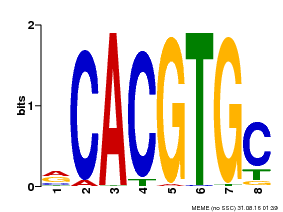
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by methyl jasmonate and wounding. {ECO:0000269|PubMed:15231736}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020094338.1 | 1e-177 | LOW QUALITY PROTEIN: transcription factor MYC2-like | ||||
| Swissprot | A0A060KY90 | 1e-145 | MYC1_SOLLC; Transcription factor MYC1 | ||||
| TrEMBL | M0S905 | 0.0 | M0S905_MUSAM; Uncharacterized protein | ||||
| STRING | GSMUA_Achr2P18140_001 | 0.0 | (Musa acuminata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP4452 | 31 | 57 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G17880.1 | 1e-127 | bHLH family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



