 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSMUA_Achr4P31770_001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Zingiberales; Musaceae; Musa
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 593aa MW: 65122.3 Da PI: 6.342 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 53.7 | 3.8e-17 | 312 | 358 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L++l+P++ K +Ka++L +A+eY+ksLq
GSMUA_Achr4P31770_001 312 VHNLSERRRRDRINEKMKALQDLIPHC-----NKTDKASMLDEAIEYLKSLQ 358
6*************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00083 | 7.55E-18 | 303 | 362 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 2.0E-21 | 305 | 366 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 3.93E-21 | 305 | 373 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.584 | 308 | 357 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.5E-14 | 312 | 358 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.3E-19 | 314 | 363 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009585 | Biological Process | red, far-red light phototransduction | ||||
| GO:0009693 | Biological Process | ethylene biosynthetic process | ||||
| GO:0010244 | Biological Process | response to low fluence blue light stimulus by blue low-fluence system | ||||
| GO:0010600 | Biological Process | regulation of auxin biosynthetic process | ||||
| GO:0010928 | Biological Process | regulation of auxin mediated signaling pathway | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 593 aa Download sequence Send to blast |
MSCGLLFVNF RIWPWLKEQR VKCLVGELRG ALSEAAIHQA MNHYVPDWAM DDDSGCLTDL 60 LPMTNQKKPM GPDNELIELL WRNGHVVMHS QSHRRPPANV DELKQAQKPD QVQKHEQQPL 120 GGSGNLMQDA DTGSWFQYPL DDSFEKEFCS EFFPEITGGA DAAADSDKIS KDLGGGSSST 180 KAGAQESSMM TVGSSTCGSN QIHAHTDPSN NLSHDAADIV TGLEEDTRMR VLSEGMQSKA 240 NECTLTSTSG GSGSSYGRTG QQNASDRSHK RKARDDVDDS GCQSEEVEYE SIEEKKPAQR 300 PISKRRSRAA EVHNLSERRR RDRINEKMKA LQDLIPHCNK TDKASMLDEA IEYLKSLQLQ 360 VQMMWMGSGM ASMMFPGVQQ YISSMGMGMG MGMGHASVPA IHGAVQLPRV PFVNQSVATA 420 SGTNQTSFFP SPAMNAVNFP NQMQNIHLPE SYARYLGMPI MPSHQATNFC TYGSQPVQQN 480 QSAGAPGGSL RPGAGGPNCA STENNRSDIS IVLCISINIL PLLHSQLHST IAFFHLVSFG 540 WISSNIFQRF LTLKLVNNSR IQGLTGDHLI CYLYLEQDWF LSAKIESGLS PVV |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 303 | 320 | KRRSRAAEVHNLSERRRR |
| 2 | 316 | 321 | ERRRRD |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00082 | ChIP-seq | Transfer from AT3G59060 | Download |
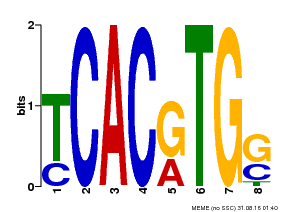
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009397370.1 | 0.0 | PREDICTED: transcription factor PIF4-like | ||||
| Refseq | XP_009397371.1 | 0.0 | PREDICTED: transcription factor PIF4-like | ||||
| TrEMBL | M0STT9 | 0.0 | M0STT9_MUSAM; Uncharacterized protein | ||||
| STRING | GSMUA_Achr4P31770_001 | 0.0 | (Musa acuminata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3107 | 36 | 77 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G59060.1 | 3e-42 | phytochrome interacting factor 3-like 6 | ||||



