 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSMUA_Achr4P33130_001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Zingiberales; Musaceae; Musa
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 368aa MW: 40115.9 Da PI: 9.548 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 67 | 3.2e-21 | 271 | 334 | 1 | 64 |
XXXXCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHC CS
bZIP_1 1 ekelkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklkseve 64
++el+re+rkq+NRe+ArrsR+RK++e+eeL+++v Le+eN+aL+ e+e+lkk+ +lk+e++
GSMUA_Achr4P33130_001 271 DRELRREKRKQSNRESARRSRLRKQQECEELARRVTDLESENSALRVEIESLKKLRGELKAENK 334
689*********************************************************9985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF07777 | 8.3E-19 | 36 | 112 | IPR012900 | G-box binding protein, multifunctional mosaic region |
| Pfam | PF16596 | 3.6E-10 | 138 | 267 | No hit | No description |
| PRINTS | PR00041 | 3.5E-6 | 181 | 203 | IPR001630 | cAMP response element binding (CREB) protein |
| Gene3D | G3DSA:1.20.5.170 | 1.4E-16 | 267 | 330 | No hit | No description |
| PRINTS | PR00041 | 3.5E-6 | 270 | 286 | IPR001630 | cAMP response element binding (CREB) protein |
| SMART | SM00338 | 1.5E-18 | 271 | 335 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 4.4E-19 | 272 | 334 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 13.185 | 273 | 336 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 1.25E-10 | 274 | 330 | No hit | No description |
| CDD | cd14702 | 1.56E-20 | 276 | 324 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 278 | 293 | IPR004827 | Basic-leucine zipper domain |
| PRINTS | PR00041 | 3.5E-6 | 288 | 308 | IPR001630 | cAMP response element binding (CREB) protein |
| PRINTS | PR00041 | 3.5E-6 | 308 | 325 | IPR001630 | cAMP response element binding (CREB) protein |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010310 | Biological Process | regulation of hydrogen peroxide metabolic process | ||||
| GO:0010629 | Biological Process | negative regulation of gene expression | ||||
| GO:0090342 | Biological Process | regulation of cell aging | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005737 | Cellular Component | cytoplasm | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 368 aa Download sequence Send to blast |
MNIEQIFKKI HLFRVFLSKC SHFRNLLTPT QEPPTPIPPA TTSVLPEWSA SLQAFYGRGA 60 AAMMPQAFYP PPPIAAPQPV VWGAQQVMPP YGSPIAYASF YPHGGFFAQP PMNAAMAYRT 120 PETQGRPPEA KDKQITSKGT SKDGGELTGK WGNGGKGTSG AAENSSQSDD SATEGSSDTR 180 EDGSQPKDHS LARKRSYGNM IAEGEASHPL DTAEHSGANA ESTYSGRSRT AKKLPVSAPG 240 RATLPGSQTN LNIGMDFWGA THAGSVPMKD DRELRREKRK QSNRESARRS RLRKQQECEE 300 LARRVTDLES ENSALRVEIE SLKKLRGELK AENKSIMEKL KQRYGPENFS ELGISIDPSK 360 LEPDGASG |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 287 | 293 | RRSRLRK |
| 2 | 287 | 294 | RRSRLRKQ |
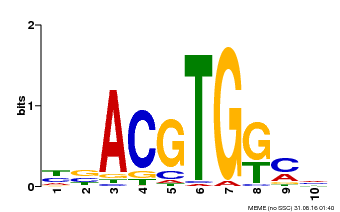
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00039 | PBM | Transfer from AT4G36730 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC186756 | 1e-102 | AC186756.1 Musa acuminata clone MA4_112I10, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009397218.1 | 0.0 | PREDICTED: G-box-binding factor 1-like | ||||
| TrEMBL | M0SU75 | 0.0 | M0SU75_MUSAM; Uncharacterized protein | ||||
| STRING | GSMUA_Achr4P33130_001 | 0.0 | (Musa acuminata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2160 | 36 | 92 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G36730.1 | 2e-40 | G-box binding factor 1 | ||||



