 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSMUA_Achr5P09950_001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Zingiberales; Musaceae; Musa
|
||||||||
| Family | RAV | ||||||||
| Protein Properties | Length: 322aa MW: 35809.5 Da PI: 9.6824 | ||||||||
| Description | RAV family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 42.6 | 1.5e-13 | 60 | 108 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
s+ykGV + +grW A+I++ ++r++lg+f ++eAa+a++ a+++++g
GSMUA_Achr5P09950_001 60 SQYKGVVPQP-NGRWGAQIYEK-----HQRVWLGTFNEEDEAARAYDVAAQRFRG 108
78****9888.8*********3.....5**********99*************98 PP
| |||||||
| 2 | B3 | 101.5 | 4.9e-32 | 146 | 247 | 1 | 96 |
EEEE-..-HHHHTT-EE--HHH.HTT......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEE CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvF 82
f+k+ tpsdv+k++rlv+pk++ae+h g +++ l +ed g++W+++++y+++s++yvltkGW++Fvk++gL++gD+v+F
GSMUA_Achr5P09950_001 146 FDKAVTPSDVGKLNRLVIPKQHAEKHfplnqtGAAACKGVLLNFEDACGKVWRFRYSYWNSSQSYVLTKGWSRFVKEKGLRAGDVVTF 233
89*************************8887755555999************************************************ PP
EE-SSSEE..EEEE CS
B3 83 kldgrsefelvvkv 96
+++ e++l++++
GSMUA_Achr5P09950_001 234 QRSTGPEKQLFIGW 247
*8866777788877 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 1.30E-24 | 60 | 115 | No hit | No description |
| SuperFamily | SSF54171 | 1.44E-17 | 60 | 116 | IPR016177 | DNA-binding domain |
| PROSITE profile | PS51032 | 20.323 | 61 | 116 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 6.4E-9 | 61 | 108 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 6.7E-21 | 61 | 116 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 4.8E-29 | 61 | 122 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 9.4E-5 | 62 | 73 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 9.4E-5 | 82 | 98 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:2.40.330.10 | 1.1E-41 | 137 | 253 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 1.05E-30 | 143 | 249 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 3.03E-28 | 144 | 236 | No hit | No description |
| PROSITE profile | PS50863 | 13.944 | 146 | 250 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 1.3E-28 | 146 | 248 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 1.4E-23 | 146 | 251 | IPR003340 | B3 DNA binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 322 aa Download sequence Send to blast |
MDTSSLEDAC SEIEKQSPPP RPRPPPPAPP KGLRRLGSGA SVVLEPEISI EAESRKLPSS 60 QYKGVVPQPN GRWGAQIYEK HQRVWLGTFN EEDEAARAYD VAAQRFRGRD AVTNFKPLVE 120 TRDEDTAELS FLNSHSKAEI HREHLFDKAV TPSDVGKLNR LVIPKQHAEK HFPLNQTGAA 180 ACKGVLLNFE DACGKVWRFR YSYWNSSQSY VLTKGWSRFV KEKGLRAGDV VTFQRSTGPE 240 KQLFIGWKTR TAGAHTADGV QATKPPVEVV RLFGVNIVKI PAAVSGDVSD RSGVGWAAKR 300 TRDMDLISSL ELFKKQCREE GS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wid_A | 2e-57 | 143 | 259 | 11 | 126 | DNA-binding protein RAV1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (By similarity). Transcriptional repressor of flowering time on long day plants. Acts directly on FT expression by binding 5'-CAACA-3' and 5'-CACCTG-3 sequences (Probable). Functionally redundant with TEM1. {ECO:0000250, ECO:0000269|PubMed:18718758, ECO:0000305}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00025 | PBM | Transfer from AT1G68840 | Download |
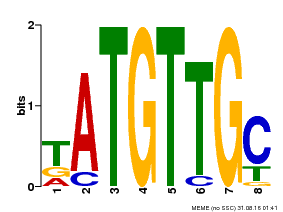
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009400502.1 | 0.0 | PREDICTED: AP2/ERF and B3 domain-containing transcription repressor RAV2-like | ||||
| Swissprot | P82280 | 1e-121 | RAV2_ARATH; AP2/ERF and B3 domain-containing transcription repressor RAV2 | ||||
| TrEMBL | M0SX74 | 0.0 | M0SX74_MUSAM; Uncharacterized protein | ||||
| STRING | GSMUA_Achr5P09950_001 | 0.0 | (Musa acuminata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP456 | 38 | 203 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G68840.2 | 1e-121 | related to ABI3/VP1 2 | ||||



