 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSMUA_Achr6P18720_001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Zingiberales; Musaceae; Musa
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 274aa MW: 30883.2 Da PI: 9.0575 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 163 | 1.1e-50 | 8 | 132 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpkkvkaeekewyfFskrdkkyatgkrknratksgyWkatgk 88
lppGfrFhPtdeelv++yL+++++g ++ + +i+e+d+yk++Pw+Lp ++ +ekewyfFs+r++ky++g+r+nra+ sgyWkatg
GSMUA_Achr6P18720_001 8 LPPGFRFHPTDEELVMHYLCRRCAGLPVAV-PIIAELDLYKFDPWHLPGMALYGEKEWYFFSPRERKYPNGSRPNRAAGSGYWKATGA 94
79*************************999.88***************6666689********************************* PP
NAM 89 dkevlskkgelvglkktLvfykgrapkgektdWvmheyrl 128
dk+v s + v +kk Lvfy+g+apkg kt+W+mheyrl
GSMUA_Achr6P18720_001 95 DKPVGS--PKPVAIKKALVFYTGKAPKGDKTNWIMHEYRL 132
****99..789***************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 1.44E-61 | 5 | 158 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 59.585 | 8 | 158 | IPR003441 | NAC domain |
| Pfam | PF02365 | 2.1E-26 | 9 | 132 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0009788 | Biological Process | negative regulation of abscisic acid-activated signaling pathway | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 274 aa Download sequence Send to blast |
MNGGDLQLPP GFRFHPTDEE LVMHYLCRRC AGLPVAVPII AELDLYKFDP WHLPGMALYG 60 EKEWYFFSPR ERKYPNGSRP NRAAGSGYWK ATGADKPVGS PKPVAIKKAL VFYTGKAPKG 120 DKTNWIMHEY RLADVDRSAR KKNSLRLDDW VLCRIYNKKG SSGSEKLGNT DGKPVVSRSV 180 WPGPATSPEE RKPVLGPYAP SPLAHVPAEM VYSFAPSDSM PKLHADSSCS ERPEFTCERE 240 VQSQPRWRPT TEWDRALGLE DPFQDILTYL GKPF |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ut4_A | 2e-76 | 5 | 162 | 14 | 169 | NO APICAL MERISTEM PROTEIN |
| 1ut4_B | 2e-76 | 5 | 162 | 14 | 169 | NO APICAL MERISTEM PROTEIN |
| 1ut7_A | 2e-76 | 5 | 162 | 14 | 169 | NO APICAL MERISTEM PROTEIN |
| 1ut7_B | 2e-76 | 5 | 162 | 14 | 169 | NO APICAL MERISTEM PROTEIN |
| 3swm_A | 2e-76 | 5 | 162 | 17 | 172 | NAC domain-containing protein 19 |
| 3swm_B | 2e-76 | 5 | 162 | 17 | 172 | NAC domain-containing protein 19 |
| 3swm_C | 2e-76 | 5 | 162 | 17 | 172 | NAC domain-containing protein 19 |
| 3swm_D | 2e-76 | 5 | 162 | 17 | 172 | NAC domain-containing protein 19 |
| 3swp_A | 2e-76 | 5 | 162 | 17 | 172 | NAC domain-containing protein 19 |
| 3swp_B | 2e-76 | 5 | 162 | 17 | 172 | NAC domain-containing protein 19 |
| 3swp_C | 2e-76 | 5 | 162 | 17 | 172 | NAC domain-containing protein 19 |
| 3swp_D | 2e-76 | 5 | 162 | 17 | 172 | NAC domain-containing protein 19 |
| 4dul_A | 2e-76 | 5 | 162 | 14 | 169 | NAC domain-containing protein 19 |
| 4dul_B | 2e-76 | 5 | 162 | 14 | 169 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the promoter of the stress response gene LEA19. Involved in tolerance to abiotic stresses (PubMed:20632034). Transcription activator involved in response to abiotic and biotic stresses. Involved in drought and salt stress responses, and defense response to the rice blast fungus (PubMed:17587305). Transcription activator involved tolerance to cold and salt stresses (PubMed:18273684). Transcription activator involved in tolerance to drought stress. Targets directly and activates genes involved in membrane modification, nicotianamine (NA) biosynthesis, glutathione relocation, accumulation of phosphoadenosine phosphosulfate and glycosylation in roots (PubMed:27892643). Controls root growth at early vegetative stage through chromatin modification and histone lysine deacytaltion by HDAC1 (PubMed:19453457). {ECO:0000269|PubMed:17587305, ECO:0000269|PubMed:18273684, ECO:0000269|PubMed:19453457, ECO:0000269|PubMed:20632034, ECO:0000269|PubMed:27892643}. | |||||
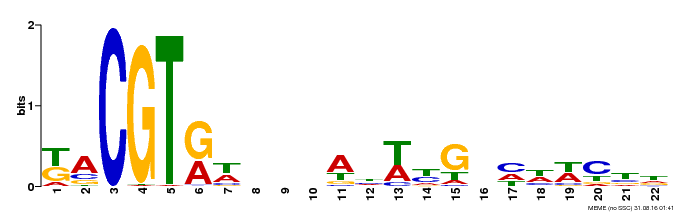
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00121 | DAP | Transfer from AT1G01720 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by drought stress, salt stress, cold stress and abscisic acid (ABA) (PubMed:20632034, PubMed:27892643). Induced by methyl jasmonate (PubMed:20632034, PubMed:11332734). Induced by infection with the rice blast fungus Magnaporthe oryzae (PubMed:11332734). {ECO:0000269|PubMed:11332734, ECO:0000269|PubMed:20632034, ECO:0000269|PubMed:27892643}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AB028185 | 1e-146 | AB028185.1 Oryza sativa mRNA for OsNAC6 protein, complete cds. | |||
| GenBank | AK068392 | 1e-146 | AK068392.1 Oryza sativa Japonica Group cDNA clone:J013149P14, full insert sequence. | |||
| GenBank | CT841730 | 1e-146 | CT841730.1 Oryza rufipogon (W1943) cDNA clone: ORW1943C102I24, full insert sequence. | |||
| GenBank | EU846993 | 1e-146 | EU846993.1 Oryza sativa Japonica Group clone KCS089G05 NAC6 protein mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009404969.2 | 0.0 | PREDICTED: NAC domain-containing protein 48-like | ||||
| Swissprot | Q7F2L3 | 1e-127 | NAC48_ORYSJ; NAC domain-containing protein 48 | ||||
| TrEMBL | M0T870 | 0.0 | M0T870_MUSAM; Uncharacterized protein | ||||
| STRING | GSMUA_Achr6P18720_001 | 0.0 | (Musa acuminata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3417 | 37 | 82 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G01720.1 | 1e-113 | NAC family protein | ||||



