 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSMUA_Achr6P31750_001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Zingiberales; Musaceae; Musa
|
||||||||
| Family | HSF | ||||||||
| Protein Properties | Length: 486aa MW: 55083.8 Da PI: 6.5179 | ||||||||
| Description | HSF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HSF_DNA-bind | 113.9 | 1.1e-35 | 14 | 105 | 2 | 102 |
HHHHHHHHHCTGGGTTTSEESSSSSEEEES-HHHHHHHTHHHHSTT--HHHHHHHHHHTTEEE---SSBTTTTXTTSEEEEESXXXXX CS
HSF_DNA-bind 2 FlkklyeiledeelkeliswsengnsfvvldeeefakkvLpkyFkhsnfaSFvRQLnmYgFkkvkdeekkskskekiweFkhksFkkg 89
Fl+k+ye+++d+++++++sws +++sfvv+++ efa+++LpkyFkh+nf+SFvRQLn+YgF+k++ ++ weF+++ F +g
GSMUA_Achr6P31750_001 14 FLTKTYEMVDDPSTNSIVSWSPSNTSFVVWNPPEFARDLLPKYFKHNNFSSFVRQLNTYGFRKTDPDQ---------WEFTNDDFIRG 92
9********************999****************************************9998.........*********** PP
XXXXXXXXXXXXX CS
HSF_DNA-bind 90 kkellekikrkks 102
+++ll++i r+k
GSMUA_Achr6P31750_001 93 QRHLLQNIYRRKP 105
**********985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.10 | 1.8E-39 | 7 | 97 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SuperFamily | SSF46785 | 4.08E-36 | 9 | 103 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SMART | SM00415 | 4.0E-65 | 10 | 103 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| Pfam | PF00447 | 8.4E-32 | 14 | 103 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 2.2E-20 | 14 | 37 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 2.2E-20 | 52 | 64 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 2.2E-20 | 65 | 77 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000302 | Biological Process | response to reactive oxygen species | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 486 aa Download sequence Send to blast |
MEGSRGSSNS PPPFLTKTYE MVDDPSTNSI VSWSPSNTSF VVWNPPEFAR DLLPKYFKHN 60 NFSSFVRQLN TYGFRKTDPD QWEFTNDDFI RGQRHLLQNI YRRKPVHSHS LHNQGNTSEA 120 ERQELEEEIE RLKQKKGTLL SEFQKQTQQR HGMEHQMQSL EDRLQVLENR QRSLMDFLAQ 180 IIQKPGFLSN IVQFSDLRSK KRRLPKIDFL GEDAIMEDNQ IASFQPVARE KSDILPMYAL 240 DMEPFEKMES SLNSLENFFK SVSQASGDDV CYDNIHSSPE VAESTSDAET SVIPATEIQT 300 DSQSKVSKID MNLAPAAIDI DSSRGQTTGT VAQTGANDVF WEQFLTEIPG SSETKEVQSK 360 RRDLDDKQSQ GEMGERGNTW CDRKNYCLFC RVQGVLKSVF VRGSPSVFGF SLGYRTRYSK 420 GCSCLLLRSA IELFVSITRY SLQIRISPHC RSTASRSIMQ SSSLAGAREG TLLGLVLLIP 480 NGHSAC |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5d5u_B | 1e-27 | 3 | 103 | 19 | 129 | Heat shock factor protein 1 |
| 5d5v_B | 1e-27 | 3 | 103 | 19 | 129 | Heat shock factor protein 1 |
| 5d5v_D | 1e-27 | 3 | 103 | 19 | 129 | Heat shock factor protein 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional regulator that specifically binds DNA of heat shock promoter elements (HSE). {ECO:0000250}. | |||||
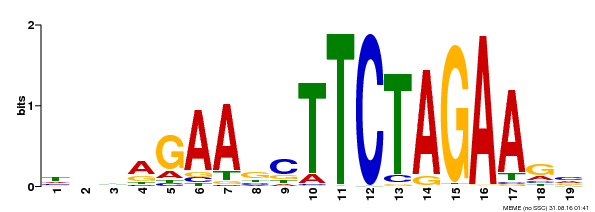
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00443 | DAP | Transfer from AT4G18880 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018683306.1 | 0.0 | PREDICTED: heat stress transcription factor A-4b-like | ||||
| Swissprot | Q94J16 | 1e-123 | HFA4B_ORYSJ; Heat stress transcription factor A-4b | ||||
| TrEMBL | M0TBX0 | 0.0 | M0TBX0_MUSAM; Uncharacterized protein | ||||
| STRING | GSMUA_Achr6P31750_001 | 0.0 | (Musa acuminata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3296 | 37 | 78 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G18880.1 | 1e-83 | heat shock transcription factor A4A | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



