 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSMUA_Achr7P04720_001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Zingiberales; Musaceae; Musa
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 469aa MW: 51253.8 Da PI: 7.6324 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 39.7 | 8.8e-13 | 312 | 356 | 5 | 54 |
HHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHH CS
HLH 5 hnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksL 54
h e+Er+RR+++N++f Lr+++P+ K++Ka+ L Av YI++L
GSMUA_Achr7P04720_001 312 HVEAERQRREKLNQRFYALRSVVPNV-----SKMDKASLLADAVTYINEL 356
899**********************6.....5***************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 1.9E-42 | 69 | 221 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| PROSITE profile | PS50888 | 16.736 | 307 | 356 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 1.83E-17 | 311 | 378 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 6.1E-18 | 312 | 372 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 5.97E-14 | 312 | 357 | No hit | No description |
| Pfam | PF00010 | 2.8E-10 | 312 | 356 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 6.4E-15 | 313 | 362 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042538 | Biological Process | hyperosmotic salinity response | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005509 | Molecular Function | calcium ion binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 469 aa Download sequence Send to blast |
MNLWNDDNTS VMEAFLSSSA TVAAPDHHSF WPPLPPPPAS IPASSSSTTS STHSTVTSSA 60 PAHFSPDTLQ QRLHSLIEGA REKWTYAIFW QSSPPSGAAV LTWGDGYYRG CEEDKRKPLR 120 SGASSAAEQE HRKHWFFLVS MTQTFAPGAG HPGQAFLSGS PAWIAGADRM AAAPCERVRQ 180 AQAFGLRTMA CVPLESGVVE LGSTHDIFQS SEILSKVRLL FGQGSGRRPA AASWPTPQPA 240 AFLSSSADVV VAEVKSEGGC GGGNLFGADS DHSDLEASAR EVTSSAVMEP PEKRPKKRGR 300 KPANGREEPL DHVEAERQRR EKLNQRFYAL RSVVPNVSKM DKASLLADAV TYINELRSKM 360 QALESDKRKL QPAASNWPGR CSEVEVEVKI LGWEAMIRVQ CDRRCHPSAR LMIALRELDL 420 EVYYANVSVV KDLMIQQVTV KMTSRIYTQE QLTAALFSSV TADHPPHAM |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4rqw_A | 3e-44 | 63 | 221 | 3 | 192 | Transcription factor MYC3 |
| 4rqw_B | 3e-44 | 63 | 221 | 3 | 192 | Transcription factor MYC3 |
| 4rs9_A | 3e-44 | 63 | 221 | 3 | 192 | Transcription factor MYC3 |
| 4yz6_A | 3e-44 | 63 | 221 | 3 | 192 | Transcription factor MYC3 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 292 | 300 | KRPKKRGRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator involved in jasmonate (JA) signaling pathway during spikelet development. Binds to the G2 region G-box (5'-CACGTG-3') of the MADS1 promoter and thus directly regulates the expression of MADS1. Its function in MADS1 activation is abolished by TIFY3/JAZ1 which directly target MYC2 during spikelet development. {ECO:0000269|PubMed:24647160}. | |||||
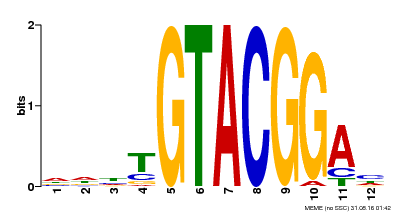
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00547 | DAP | Transfer from AT5G46830 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009408104.1 | 1e-145 | PREDICTED: transcription factor MYC2-like | ||||
| Swissprot | Q336P5 | 1e-88 | MYC2_ORYSJ; Transcription factor MYC2 | ||||
| TrEMBL | M0TER2 | 0.0 | M0TER2_MUSAM; Uncharacterized protein | ||||
| STRING | GSMUA_Achr7P04720_001 | 0.0 | (Musa acuminata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP4452 | 31 | 57 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G46830.1 | 7e-89 | NACL-inducible gene 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



