 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSMUA_Achr7P11600_001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Zingiberales; Musaceae; Musa
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 276aa MW: 31647.3 Da PI: 6.5108 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 57.6 | 2.2e-18 | 75 | 128 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k++++t eq+++Le+ Fe +++ e++ +LA+ lg++ rqV +WFqNrRa++k
GSMUA_Achr7P11600_001 75 KKRRLTVEQVRTLEKNFELGNKLEPERKMQLARTLGMQPRQVAIWFQNRRARWK 128
45689************************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 131.2 | 4e-42 | 74 | 166 | 1 | 93 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLr 88
ekkrrl+ eqv++LE++Fe +kLeperK++lar+Lg+qprqva+WFqnrRAR+ktkqlEkdy++Lkr+++a+k+ene+L++++++L+
GSMUA_Achr7P11600_001 74 EKKRRLTVEQVRTLEKNFELGNKLEPERKMQLARTLGMQPRQVAIWFQNRRARWKTKQLEKDYDVLKRHFEAIKSENEALKAHNNQLQ 161
69************************************************************************************** PP
HD-ZIP_I/II 89 eelke 93
+e+++
GSMUA_Achr7P11600_001 162 AEIMT 166
99975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 5.56E-20 | 65 | 132 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.41 | 70 | 130 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 5.4E-18 | 73 | 134 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 9.6E-16 | 75 | 128 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 7.12E-16 | 75 | 131 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 1.5E-19 | 77 | 137 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 1.8E-5 | 101 | 110 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 105 | 128 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 1.8E-5 | 110 | 126 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 1.7E-16 | 130 | 170 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009744 | Biological Process | response to sucrose | ||||
| GO:0048826 | Biological Process | cotyledon morphogenesis | ||||
| GO:0080022 | Biological Process | primary root development | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 276 aa Download sequence Send to blast |
MASSFFHHGF MLQMQTPHEE EHQLLTPSPC TNLQDLRGTA SMVGKRSVSF SGMDTCREMS 60 AEEDLSDDGS QAGEKKRRLT VEQVRTLEKN FELGNKLEPE RKMQLARTLG MQPRQVAIWF 120 QNRRARWKTK QLEKDYDVLK RHFEAIKSEN EALKAHNNQL QAEIMTLRGR EPSDLINLNK 180 DTEGSCSNRS ENSSEINLDI SRQSVDKSPL HPHQSSTFFQ PVRPGDMDQI FQSSSRPETQ 240 CPKLENGGHE GTLSNLLCSM EDQSAFWSWS DHHSFH |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 122 | 130 | RRARWKTKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00225 | DAP | Transfer from AT1G69780 | Download |
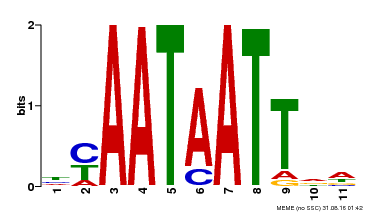
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | JX620522 | 4e-88 | JX620522.1 Gossypium hirsutum clone NBRI_GE66779 microsatellite sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009409146.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein HOX21-like | ||||
| Swissprot | Q8S7W9 | 1e-92 | HOX21_ORYSJ; Homeobox-leucine zipper protein HOX21 | ||||
| TrEMBL | M0TGP8 | 0.0 | M0TGP8_MUSAM; Uncharacterized protein | ||||
| STRING | GSMUA_Achr7P11600_001 | 0.0 | (Musa acuminata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2289 | 38 | 94 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G69780.1 | 2e-93 | HD-ZIP family protein | ||||



