 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSMUA_Achr7P20100_001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Zingiberales; Musaceae; Musa
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 358aa MW: 39583.9 Da PI: 4.9063 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 13.6 | 0.00013 | 54 | 71 | 2 | 19 |
HHHHHHHHHHHHHHHHHH CS
HLH 2 rrahnerErrRRdriNsa 19
r +h+++E+rRR++iN++
GSMUA_Achr7P20100_001 54 RSKHSAMEQRRRNKINDR 71
899*************96 PP
| |||||||
| 2 | HLH | 25.5 | 2.4e-08 | 87 | 118 | 20 | 55 |
HHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 20 feeLrellPkaskapskKlsKaeiLekAveYIksLq 55
f+ Lrel+P++ ++K +Ka+ L +++eYI+ Lq
GSMUA_Achr7P20100_001 87 FQILRELIPHS----DQKRDKASFLLEVIEYIQFLQ 118
89********9....9*******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.280.10 | 8.5E-21 | 52 | 141 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 11.287 | 52 | 117 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 5.37E-13 | 52 | 142 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 2.50E-14 | 54 | 122 | No hit | No description |
| SMART | SM00353 | 2.3E-6 | 58 | 123 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.1E-5 | 87 | 118 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006351 | Biological Process | transcription, DNA-templated | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 358 aa Download sequence Send to blast |
MEAAASRSGR GFDDEDDDDQ EFAKREGFFF SHKDLTIKAD GKGSGTDDLP TTPRSKHSAM 60 EQRRRNKIND RQARPPAFLM EDKFCLFQIL RELIPHSDQK RDKASFLLEV IEYIQFLQEK 120 VQKYESSYPG WNQDNAKLMP WVKVYYRSFW KNAQNNNQTP VDGLSDPSQV IRNGSAPPAS 180 VFAGQFDEND IPAAPAMLSN APNPTQLDMT TGVPYKIMET ATGFAIADNM PSQAQPHWLG 240 SSSPADCAVS SEMLNEQEEL IIDEGTINAS ASYSQGLLTS LTQTLQSSGV DLSQANISVQ 300 INLGKRATNR RPTATTALSN YKDPDDPAST NQAVGRSMMG ITNQESSQAA KRHKADNC |
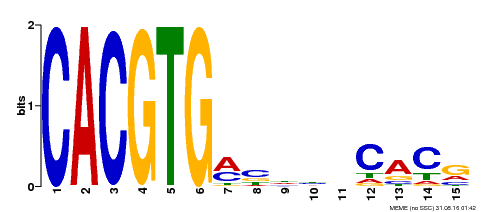
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00219 | DAP | Transfer from AT1G69010 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009410295.1 | 0.0 | PREDICTED: transcription factor BIM2 isoform X1 | ||||
| TrEMBL | M0TJ47 | 0.0 | M0TJ47_MUSAM; Uncharacterized protein | ||||
| STRING | GSMUA_Achr7P20100_001 | 0.0 | (Musa acuminata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP762 | 37 | 128 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G69010.1 | 3e-51 | BES1-interacting Myc-like protein 2 | ||||



