 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSMUA_Achr7P27540_001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Zingiberales; Musaceae; Musa
|
||||||||
| Family | TALE | ||||||||
| Protein Properties | Length: 350aa MW: 38876.9 Da PI: 5.6196 | ||||||||
| Description | TALE family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 26.9 | 8.5e-09 | 273 | 311 | 17 | 55 |
HHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHH CS
Homeobox 17 elFeknrypsaeereeLAkklgLterqVkvWFqNrRake 55
el +k +yps++++ LA+++gL+ +q+ +WF N+R ++
GSMUA_Achr7P27540_001 273 ELHYKWPYPSETQKVALAESTGLDLKQINNWFINQRKRH 311
5556789*****************************885 PP
| |||||||
| 2 | ELK | 39.7 | 9.9e-14 | 231 | 252 | 1 | 22 |
ELK 1 ELKhqLlrKYsgyLgsLkqEFs 22
ELKhqLlrKYsgyL+sL+qE+s
GSMUA_Achr7P27540_001 231 ELKHQLLRKYSGYLSSLRQELS 252
9*******************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01255 | 7.0E-22 | 86 | 130 | IPR005540 | KNOX1 |
| Pfam | PF03790 | 3.2E-24 | 87 | 129 | IPR005540 | KNOX1 |
| SMART | SM01256 | 2.0E-27 | 137 | 188 | IPR005541 | KNOX2 |
| Pfam | PF03791 | 1.0E-23 | 143 | 186 | IPR005541 | KNOX2 |
| SMART | SM01188 | 1800 | 146 | 166 | IPR005539 | ELK domain |
| Pfam | PF03789 | 1.2E-10 | 231 | 252 | IPR005539 | ELK domain |
| PROSITE profile | PS51213 | 11.592 | 231 | 251 | IPR005539 | ELK domain |
| SMART | SM01188 | 7.2E-8 | 231 | 252 | IPR005539 | ELK domain |
| PROSITE profile | PS50071 | 12.536 | 251 | 314 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 1.37E-19 | 252 | 327 | IPR009057 | Homeodomain-like |
| SMART | SM00389 | 2.0E-12 | 253 | 318 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 1.4E-27 | 256 | 316 | IPR009057 | Homeodomain-like |
| CDD | cd00086 | 1.71E-12 | 263 | 315 | No hit | No description |
| Pfam | PF05920 | 3.8E-16 | 271 | 310 | IPR008422 | Homeobox KN domain |
| PROSITE pattern | PS00027 | 0 | 289 | 312 | IPR017970 | Homeobox, conserved site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0001708 | Biological Process | cell fate specification | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010051 | Biological Process | xylem and phloem pattern formation | ||||
| GO:0010089 | Biological Process | xylem development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 350 aa Download sequence Send to blast |
MEEFSQLGGN LWGGLMCSTS PAAKSTGDTG AVAMVAPPSG GASVFDKGHG CIPNPLLLGS 60 GGNQNSHLLA KHEGASSSHF PPCDVDAIKA KIISHPQYSS LLAAYIDCQK VGAPPEVVDR 120 LSAVAQELEM RQRASLICRD APTDPELDQF MEAYHEMLVK YREELTRPLQ EAMEFLRRVE 180 AQLNSLSLTD GSLRILSADD KFGVCSSEDD QDGSGGETEL PELDASAEDR ELKHQLLRKY 240 SGYLSSLRQE LSKKKKKGKL PKDARQQLLN WWELHYKWPY PSETQKVALA ESTGLDLKQI 300 NNWFINQRKR HWKPSEDMQF VVMDGYHAPN AALYMDGQYF MGDGLYRLGP |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | May play a role in meristem formation and/or maintenance. Overexpression causes the hooded phenotype characterized by the appearance of an extra flower of inverse polarity on the lemma. Binds to the DNA sequence 5'-TGAC-3'. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00675 | ChIP-seq | Transfer from GRMZM2G017087 | Download |
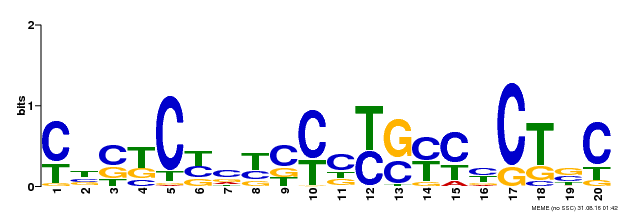
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009411331.2 | 0.0 | PREDICTED: homeobox protein knotted-1-like 6 | ||||
| Swissprot | Q43484 | 1e-143 | KNOX3_HORVU; Homeobox protein KNOX3 | ||||
| TrEMBL | M0TL90 | 0.0 | M0TL90_MUSAM; Uncharacterized protein | ||||
| STRING | GSMUA_Achr7P27540_001 | 0.0 | (Musa acuminata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP9886 | 31 | 45 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G08150.1 | 1e-117 | KNOTTED-like from Arabidopsis thaliana | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



