 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSMUA_Achr8P11070_001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Zingiberales; Musaceae; Musa
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 845aa MW: 91995.7 Da PI: 8.029 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 47.9 | 2.4e-15 | 277 | 322 | 5 | 55 |
HHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 5 hnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn+ Er+RRdriN+++ +L++l+P++ K +Ka++L +++eY+k+Lq
GSMUA_Achr8P11070_001 277 HNQSERKRRDRINQRMKTLQKLVPNS-----SKTDKASMLDEVIEYLKQLQ 322
9************************8.....7******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.280.10 | 5.2E-18 | 271 | 343 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 17.803 | 272 | 321 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 1.31E-19 | 275 | 346 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 6.1E-13 | 277 | 322 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 8.11E-17 | 277 | 326 | No hit | No description |
| SMART | SM00353 | 3.0E-18 | 278 | 327 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF13912 | 0.0056 | 454 | 476 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 5.3 | 454 | 476 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 8.517 | 454 | 476 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 456 | 476 | IPR007087 | Zinc finger, C2H2 |
| Pfam | PF13912 | 5.2E-9 | 612 | 637 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 1.15E-7 | 612 | 635 | No hit | No description |
| PROSITE profile | PS50157 | 9.827 | 613 | 640 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.13 | 613 | 635 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 615 | 635 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 1.15E-7 | 664 | 691 | No hit | No description |
| Pfam | PF13912 | 7.4E-13 | 668 | 692 | IPR007087 | Zinc finger, C2H2 |
| PROSITE profile | PS50157 | 9.349 | 669 | 691 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.46 | 669 | 691 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 671 | 691 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009567 | Biological Process | double fertilization forming a zygote and endosperm | ||||
| GO:0009506 | Cellular Component | plasmodesma | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 845 aa Download sequence Send to blast |
MSQRVPSWDV DDPPFNPSSS NLPLHRYHPS GAPLGRLMGY EVAELAWENG QLALHGLVPS 60 RTGAKPGADA ACSKHHLHHH PSGTLESVVN QATGAAARSP VLEGWLRRST SVAVDALVPC 120 QDDVASNRIA DPDAPPCCKR ARMVGVCSSQ GSAAGSLPGR VDSTTFVTLD TWREDDVGLT 180 ATATTATTAT ATDTSAWPET ENTSLGKRKV RALYDHVSIS HTSSQTKVIL MHTSTYHNTT 240 TKKKKNNNYY YYLDQNPSWE VGKPSGATKK SRAAAIHNQS ERKRRDRINQ RMKTLQKLVP 300 NSSKTDKASM LDEVIEYLKQ LQAQVQMMSR MSSMMVAAMQ QLQMPMVAQL LPHMAQFPQL 360 PHMGLMDFAS IGRSVPPVLP LLHPSAFLHL AATGGWDGIG DRRPVGSVLP DPFSALLACQ 420 MAQQPMSLDE YSRMVTLLQH LSQNQSPANL KSSHYCRICK KGFGCGRALG GHMRAHGIVD 480 DYAADAEDDP SGCSDWDGRM NSAAAGTKRM YALRTNPARL RSCREEEEEE EEGYESVPAS 540 SRSEGDVDLA GWSKGKRSRR AKVVGMSEEE DLASCLMMLS AARQTVAVGV TTEIPQGPAF 600 LPPAQPSVPR GTFECKACKK VFSSHQALGG HRASHKKNVA AASTSTAIVP FDDPAPLAIT 660 PLRKRSKVHE CSICHRVFTS GQALGGHKRC HWITSSSPDP GLKLQPLPHH ANLPHQLTLR 720 PMFDEPLDLN QPARADEIAR GTRDIGSPLR LQMPAAIYLQ AWIDRRDVGR NRACATSSDK 780 NDDHNNVHGK DDNNTEMSGL NVDDEVDSNV KRAKLSELKD INMGEDSSPW LQVGIGSSAN 840 ESSEA |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 280 | 285 | ERKRRD |
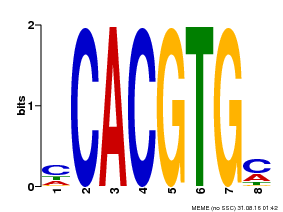
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00034 | PBM | Transfer from AT4G00050 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009411572.2 | 0.0 | PREDICTED: transcription factor UNE10 | ||||
| TrEMBL | M0TPG0 | 0.0 | M0TPG0_MUSAM; Uncharacterized protein | ||||
| STRING | GSMUA_Achr8P11070_001 | 0.0 | (Musa acuminata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3524 | 32 | 62 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00050.1 | 1e-47 | bHLH family protein | ||||



