 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSMUA_Achr8P11580_001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Zingiberales; Musaceae; Musa
|
||||||||
| Family | YABBY | ||||||||
| Protein Properties | Length: 219aa MW: 23908.4 Da PI: 7.8077 | ||||||||
| Description | YABBY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | YABBY | 231 | 2.9e-71 | 17 | 180 | 6 | 170 |
YABBY 6 sseqvCyvqCnfCntilavsvPstslfkvvtvrCGhCtsllsvnlakasqllaaeshldeslkeelleelkveeenlksnvekeesas 93
seq+Cyv+CnfC+t+lavsvP+tslfk+vtvrCGhCt+llsvn++ a + hl+++ + +++l ++ + + s
GSMUA_Achr8P11580_001 17 PSEQLCYVHCNFCDTVLAVSVPYTSLFKTVTVRCGHCTNLLSVNMRGLLLPAANQLHLSHACLTPAHHNLLDDLQCPPPSLLLDPSIL 104
68****************************************9999988888888888998888866666665555444444433333 PP
YABBY 94 tsvsseklsenedeevprvppvirPPekrqrvPsaynrfikeeiqrikasnPdishreafsaaaknWahfPkihfgl 170
+ ++ + +e +pr p v+rPPekrqrvPsaynrfik+eiqrika+nPdi+hreafsaaaknWahfP+ihfgl
GSMUA_Achr8P11580_001 105 NCSTTAP-VKGVEEVLPRSPVVNRPPEKRQRVPSAYNRFIKDEIQRIKAGNPDITHREAFSAAAKNWAHFPHIHFGL 180
3333333.46778999***********************************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04690 | 2.1E-70 | 17 | 180 | IPR006780 | YABBY protein |
| SuperFamily | SSF47095 | 1.06E-7 | 124 | 173 | IPR009071 | High mobility group box domain |
| Gene3D | G3DSA:1.10.30.10 | 2.7E-4 | 129 | 172 | IPR009071 | High mobility group box domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009909 | Biological Process | regulation of flower development | ||||
| GO:0009933 | Biological Process | meristem structural organization | ||||
| GO:0009944 | Biological Process | polarity specification of adaxial/abaxial axis | ||||
| GO:0010093 | Biological Process | specification of floral organ identity | ||||
| GO:0010154 | Biological Process | fruit development | ||||
| GO:0010158 | Biological Process | abaxial cell fate specification | ||||
| GO:0010159 | Biological Process | specification of organ position | ||||
| GO:0010450 | Biological Process | inflorescence meristem growth | ||||
| GO:1902183 | Biological Process | regulation of shoot apical meristem development | ||||
| GO:2000024 | Biological Process | regulation of leaf development | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 219 aa Download sequence Send to blast |
MSSSSAAFSL DHLSPPPSEQ LCYVHCNFCD TVLAVSVPYT SLFKTVTVRC GHCTNLLSVN 60 MRGLLLPAAN QLHLSHACLT PAHHNLLDDL QCPPPSLLLD PSILNCSTTA PVKGVEEVLP 120 RSPVVNRPPE KRQRVPSAYN RFIKDEIQRI KAGNPDITHR EAFSAAAKNW AHFPHIHFGL 180 MPDQGLKKAS VRQAPEGEDV LPKEGFYATA AANMGVTPF |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | May be involved in leaf cell growth and differentiation, rather than abaxial cell fate determination. {ECO:0000250}. | |||||
| UniProt | May be involved in leaf cell growth and differentiation, rather than abaxial cell fate determination. {ECO:0000269|PubMed:17351053}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00620 | PBM | Transfer from AT2G45190 | Download |
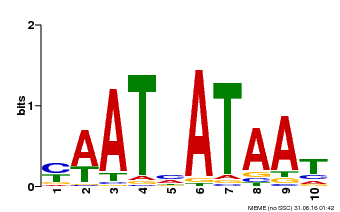
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by WOX3. {ECO:0000269|PubMed:17351053}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018685706.1 | 1e-157 | PREDICTED: protein YABBY 4-like isoform X1 | ||||
| Swissprot | Q01JG2 | 2e-84 | YAB5_ORYSI; Protein YABBY 5 | ||||
| Swissprot | Q0JBF0 | 2e-84 | YAB5_ORYSJ; Protein YABBY 5 | ||||
| TrEMBL | M0TPL1 | 1e-161 | M0TPL1_MUSAM; Uncharacterized protein | ||||
| STRING | GSMUA_Achr8P11580_001 | 1e-162 | (Musa acuminata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1445 | 38 | 121 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G45190.1 | 4e-83 | YABBY family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



