 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSMUA_Achr8P21030_001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Zingiberales; Musaceae; Musa
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 502aa MW: 55372.8 Da PI: 5.8964 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 50.4 | 5.6e-16 | 164 | 213 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV++++++grW+++I+d + k+++lg f ta Aa+a+++a+ k++g
GSMUA_Achr8P21030_001 164 SQYRGVTFYRRTGRWESHIWD------CgKQVYLGGFNTAHVAARAYDRAAIKFRG 213
78*******************......55************************997 PP
| |||||||
| 2 | AP2 | 43.3 | 9.3e-14 | 256 | 306 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpseng...krkrfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV+ +k grW+A+ g +k+++lg f+++eeAa a+++a++k +g
GSMUA_Achr8P21030_001 256 SKYRGVTLHK-CGRWEAR---M---GqllGKKYIYLGLFDSEEEAAMAYDKAALKCNG 306
89********.7******...5...334336**********************99876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF54171 | 2.75E-15 | 164 | 222 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 2.9E-8 | 164 | 213 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 2.0E-16 | 165 | 222 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 7.18E-23 | 165 | 223 | No hit | No description |
| SMART | SM00380 | 1.3E-30 | 165 | 227 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 17.187 | 165 | 221 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 1.05E-17 | 256 | 315 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 1.3E-9 | 256 | 306 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 7.59E-27 | 256 | 316 | No hit | No description |
| Gene3D | G3DSA:3.30.730.10 | 3.2E-16 | 257 | 314 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 2.4E-33 | 257 | 320 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 15.975 | 257 | 314 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 5.2E-6 | 258 | 269 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 5.2E-6 | 296 | 316 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 502 aa Download sequence Send to blast |
MLDLNVASSG ESTAVEEETI GRGTGGGGLA IGSDTSESSV LNAEASADAA ADENSCSTRP 60 VTAPAIEFGI LRSSASEEGE NDVDEENEEV RNVVSQEPAL ITRQLFPPEP PLEPFPAASS 120 SPSSSKLQWM DPSFYQADIA PQATQQQQQQ QVKKSRRGPR SRSSQYRGVT FYRRTGRWES 180 HIWDCGKQVY LGGFNTAHVA ARAYDRAAIK FRGIDADINF NLSDYEEDMK QMKNLTKEEF 240 VHILRRQSTG FSRGSSKYRG VTLHKCGRWE ARMGQLLGKK YIYLGLFDSE EEAAMAYDKA 300 ALKCNGREAV TNFDPSSYEG HLSTEADSEG QSVDLNLKIS QPDVHSPKRD QNPPEILYSY 360 SPSEASATKR VQISSPPLQP TSTPYHMDML PDKSHAWTAQ NPPFYSTTEE RDRKKRPMVG 420 LEALPNWTWQ MHGSTPLPLF SCAASSGFPT AIITAAIPPP KPPASTSSPF PSTIKLLCLT 480 TCRFFYMCKS CSLKISISIQ MD |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor (By similarity). Involved in spikelet transition. Regulator of starch biosynthesis especially during seed development (e.g. endosperm starch granules); represses the expression of type I starch synthesis genes. Prevents lemma and palea elongation as well as grain growth (By similarity). {ECO:0000250|UniProtKB:P47927, ECO:0000250|UniProtKB:Q2TQ34}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00615 | PBM | Transfer from AT2G28550 | Download |
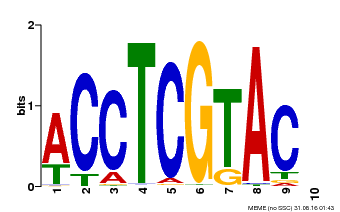
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC226052 | 0.0 | AC226052.1 Musa balbisiana clone BAC MBP-64B17, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009414295.1 | 0.0 | PREDICTED: ethylene-responsive transcription factor RAP2-7-like isoform X1 | ||||
| Refseq | XP_009414296.1 | 0.0 | PREDICTED: ethylene-responsive transcription factor RAP2-7-like isoform X1 | ||||
| Swissprot | B8AXC3 | 1e-125 | AP21_ORYSI; APETALA2-like protein 1 | ||||
| TrEMBL | M0TSA6 | 0.0 | M0TSA6_MUSAM; Uncharacterized protein | ||||
| STRING | GSMUA_Achr8P21030_001 | 0.0 | (Musa acuminata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2448 | 38 | 90 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G28550.3 | 1e-100 | related to AP2.7 | ||||



