 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSMUA_Achr8P25550_001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Zingiberales; Musaceae; Musa
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 344aa MW: 36962.9 Da PI: 8.1493 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 137.8 | 3.1e-43 | 51 | 126 | 1 | 76 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkk 76
+CqvegC+adl+ ak y+ rhkvC +hskap+v+v+gleqrfCqqCsrfh+l efD++krsCrrrLa+hnerrrk+
GSMUA_Achr8P25550_001 51 RCQVEGCNADLTGAKAYYCRHKVCGMHSKAPKVIVAGLEQRFCQQCSRFHQLPEFDQGKRSCRRRLAGHNERRRKP 126
6*************************************************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 7.7E-36 | 44 | 113 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.479 | 49 | 126 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 8.89E-41 | 50 | 130 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 3.0E-33 | 52 | 125 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0048653 | Biological Process | anther development | ||||
| GO:2000025 | Biological Process | regulation of leaf formation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 344 aa Download sequence Send to blast |
MDSSSLKASG ASSGSGDALH GLKFGQKIYF EDGGGGKGKG VAQGGQQQPP RCQVEGCNAD 60 LTGAKAYYCR HKVCGMHSKA PKVIVAGLEQ RFCQQCSRFH QLPEFDQGKR SCRRRLAGHN 120 ERRRKPPPGP FGTRYGRIAS SFHEDPNRFR SFLVDFSYPN FSCAARDVWP TARTGDRMAN 180 NEWRGVLDAP SHAAALHGTH PCFQGPPVGA FCSPMELPPG ECLAGVSDSS CALSLLSTDP 240 WSSNSAGNRS PVIPASSTFD GQPTTHSVIP SNYFNSSWGT RGHGARISSH EIQHEMGLAG 300 GTTNVSDAHF TGQVEFAPQE NGHCLDHGSD RAYDHSGPGM QWSL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 6e-31 | 52 | 125 | 11 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' (By similarity). May be involved in panicle development. {ECO:0000250}. | |||||
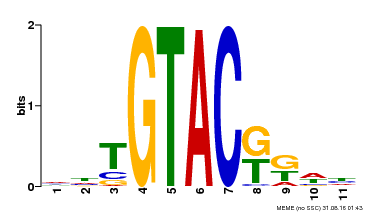
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00307 | DAP | Transfer from AT2G42200 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Negatively regulated by microRNAs miR156b and miR156h. {ECO:0000305|PubMed:16861571}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009415426.1 | 0.0 | PREDICTED: squamosa promoter-binding-like protein 14 isoform X1 | ||||
| Swissprot | A3C057 | 2e-91 | SPL17_ORYSJ; Squamosa promoter-binding-like protein 17 | ||||
| TrEMBL | M0TTK8 | 0.0 | M0TTK8_MUSAM; Uncharacterized protein | ||||
| STRING | GSMUA_Achr8P25550_001 | 0.0 | (Musa acuminata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2527 | 37 | 87 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G42200.1 | 5e-53 | squamosa promoter binding protein-like 9 | ||||



