 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSMUA_Achr8P26480_001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Zingiberales; Musaceae; Musa
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 281aa MW: 30929.9 Da PI: 8.6518 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 57.4 | 2.4e-18 | 133 | 187 | 2 | 56 |
T--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 2 rkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
rk+ ++tkeq Le+ F+++++++ +++ LAk+l+L rqV vWFqNrRa+ k
GSMUA_Achr8P26480_001 133 RKKLRLTKEQSALLEDRFKEHSTLNPKQKHALAKQLNLRPRQVEVWFQNRRARTK 187
788899***********************************************98 PP
| |||||||
| 2 | HD-ZIP_I/II | 125 | 3.5e-40 | 133 | 222 | 1 | 91 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLr 88
+kk+rl+keq++lLE+ F+e+++L+p++K++la++L+l+prqv+vWFqnrRARtk+kq+E+d+e+Lkr++++l++en+rL+ke +eL+
GSMUA_Achr8P26480_001 133 RKKLRLTKEQSALLEDRFKEHSTLNPKQKHALAKQLNLRPRQVEVWFQNRRARTKLKQTEVDCEFLKRCCESLTDENRRLQKELQELK 220
69*************************************************************************************9 PP
HD-ZIP_I/II 89 eel 91
+l
GSMUA_Achr8P26480_001 221 -AL 222
.55 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 7.7E-18 | 120 | 183 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 1.09E-18 | 124 | 190 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.184 | 129 | 189 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 4.4E-15 | 131 | 193 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 1.1E-15 | 133 | 187 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 3.07E-16 | 133 | 190 | No hit | No description |
| PROSITE pattern | PS00027 | 0 | 164 | 187 | IPR017970 | Homeobox, conserved site |
| Pfam | PF02183 | 2.0E-11 | 189 | 223 | IPR003106 | Leucine zipper, homeobox-associated |
| SMART | SM00340 | 1.5E-25 | 189 | 232 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009735 | Biological Process | response to cytokinin | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 281 aa Download sequence Send to blast |
MSPEIDACDT GLALGLGIGR RSTPSSANSR RLQEACSPLR SSMPQKQPSL TLSLSDDVDG 60 SATLLRAEAN KPSEEGRQAR PSSPHSTVSS FSAAYGPTIK KEKDVTEADE EAEVERVDSR 120 GGTCDDDNDG SARKKLRLTK EQSALLEDRF KEHSTLNPKQ KHALAKQLNL RPRQVEVWFQ 180 NRRARTKLKQ TEVDCEFLKR CCESLTDENR RLQKELQELK ALKFAPSVYM QFPAAGLTMC 240 PSCKTIGAAA DVGPKGGRAE SFVVAPPKPH FFNPFIHSAK C |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 181 | 189 | RRARTKLKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00477 | DAP | Transfer from AT4G37790 | Download |
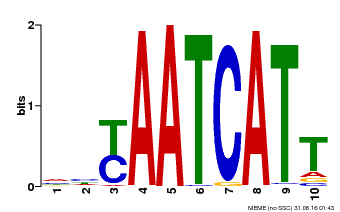
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: In leaves by drought stress. {ECO:0000269|PubMed:17999151}. | |||||
| UniProt | INDUCTION: In leaves by drought stress. {ECO:0000269|PubMed:17999151}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009415318.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein HOX19-like | ||||
| Swissprot | A2XE76 | 3e-77 | HOX19_ORYSI; Homeobox-leucine zipper protein HOX19 | ||||
| Swissprot | Q8GRL4 | 3e-77 | HOX19_ORYSJ; Homeobox-leucine zipper protein HOX19 | ||||
| TrEMBL | M0TTV1 | 0.0 | M0TTV1_MUSAM; Uncharacterized protein | ||||
| STRING | GSMUA_Achr8P26480_001 | 0.0 | (Musa acuminata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2055 | 36 | 98 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G37790.1 | 1e-64 | HD-ZIP family protein | ||||



