 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSMUA_Achr9P14570_001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Zingiberales; Musaceae; Musa
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 263aa MW: 29479.7 Da PI: 5.4009 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 60.9 | 1.9e-19 | 71 | 124 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k++++ ++q++ Le+ Fe ++++ e++ eLA+klgL+ rqV vWFqNrRa++k
GSMUA_Achr9P14570_001 71 KKRRLMPDQVHLLERSFEAENKLEPERKSELARKLGLQPRQVAVWFQNRRARWK 124
5567899**********************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 131.8 | 2.6e-42 | 70 | 161 | 1 | 92 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLr 88
ekkrrl +qv+lLE+sFe+e+kLeperK+elar+LglqprqvavWFqnrRAR+ktkqlE+d+++Lk++y++l +++++L ke++ Lr
GSMUA_Achr9P14570_001 70 EKKRRLMPDQVHLLERSFEAENKLEPERKSELARKLGLQPRQVAVWFQNRRARWKTKQLEHDFDRLKSSYNSLLSDHDSLLKENDGLR 157
69************************************************************************************** PP
HD-ZIP_I/II 89 eelk 92
+++
GSMUA_Achr9P14570_001 158 SQVI 161
9985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50071 | 17.329 | 66 | 126 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 3.1E-18 | 69 | 130 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 2.39E-17 | 71 | 127 | No hit | No description |
| SuperFamily | SSF46689 | 3.63E-18 | 71 | 137 | IPR009057 | Homeodomain-like |
| Pfam | PF00046 | 1.2E-16 | 71 | 124 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 1.0E-20 | 73 | 133 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 1.1E-6 | 97 | 106 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 101 | 124 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 1.1E-6 | 106 | 122 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 1.5E-17 | 126 | 168 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0001558 | Biological Process | regulation of cell growth | ||||
| GO:0009637 | Biological Process | response to blue light | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 263 aa Download sequence Send to blast |
MDPARLIFDS SSSHRHHGHR MLLLGSGGVS PVFGGAISAL AEGANKRRPF FTSPDEILEE 60 EYYYDEQLPE KKRRLMPDQV HLLERSFEAE NKLEPERKSE LARKLGLQPR QVAVWFQNRR 120 ARWKTKQLEH DFDRLKSSYN SLLSDHDSLL KENDGLRSQV ISLTEKLQSK AAALMAAAAS 180 VNDPASLGVP KKAEDRLSTG SGGSAVVDAN QLLHSSEESC LPDGYHSMGL VDGRVHSEED 240 DASDEGCNYY SEEDAQLGWW VWN |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 118 | 126 | RRARWKTKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00323 | DAP | Transfer from AT3G01470 | Download |
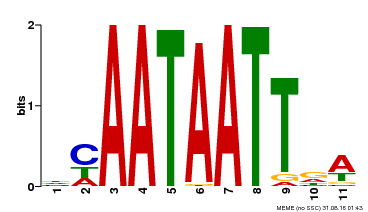
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009417556.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein HOX16-like | ||||
| Swissprot | Q6YWR4 | 1e-72 | HOX16_ORYSJ; Homeobox-leucine zipper protein HOX16 | ||||
| TrEMBL | M0U0B5 | 0.0 | M0U0B5_MUSAM; Uncharacterized protein | ||||
| STRING | GSMUA_Achr9P14570_001 | 0.0 | (Musa acuminata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2609 | 38 | 87 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G01470.1 | 3e-60 | homeobox 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



