 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSVIVT01004860001 | ||||||||
| Common Name | VIT_07s0141g00250 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; rosids incertae sedis; Vitales; Vitaceae; Vitis
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1244aa MW: 140302 Da PI: 6.351 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 142.3 | 1.4e-44 | 223 | 307 | 34 | 118 |
CG-1 34 pksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfqrrcywlLeeelekivlvhylevk 118
sgsl+L++rk++ryfrkDG++w+kkkdgktv+E+he+LK g+++vl+cyYah+e+n++fqrr+yw+Leeel++ivlvhy+evk
GSVIVT01004860001 223 LASGSLFLFDRKVLRYFRKDGHNWRKKKDGKTVKEAHERLKAGSIDVLHCYYAHGEDNENFQRRSYWMLEEELSHIVLVHYREVK 307
579*******************************************************************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 62.252 | 184 | 312 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 3.9E-51 | 188 | 307 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 8.5E-37 | 224 | 305 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 1.3E-4 | 663 | 750 | IPR013783 | Immunoglobulin-like fold |
| SuperFamily | SSF81296 | 9.62E-18 | 664 | 750 | IPR014756 | Immunoglobulin E-set |
| Pfam | PF01833 | 6.5E-7 | 664 | 749 | IPR002909 | IPT domain |
| Gene3D | G3DSA:1.25.40.20 | 1.5E-18 | 844 | 960 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 1.46E-13 | 847 | 956 | No hit | No description |
| PROSITE profile | PS50297 | 18.218 | 847 | 968 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 2.49E-18 | 849 | 958 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 1.9E-7 | 869 | 960 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 0.022 | 897 | 926 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 9.885 | 897 | 929 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 210 | 936 | 965 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 1.65E-7 | 1063 | 1118 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 17 | 1067 | 1089 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.078 | 1069 | 1088 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.071 | 1069 | 1097 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0039 | 1090 | 1112 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.597 | 1091 | 1115 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 2.6E-4 | 1093 | 1112 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045944 | Biological Process | positive regulation of transcription from RNA polymerase II promoter | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0001077 | Molecular Function | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1244 aa Download sequence Send to blast |
MPKSVARRLE KLQRDFLWEG ANGGKKAHLV KWEVVCADKE KGGLGLRKLA CLNKALLGKW 60 IWRFARAKED IWKKVLEAKY GQEDFGWRTR KANGVFGVGV WKEILKEFAW CWENMVFKVG 120 KGNKIRFWID PWCGNNVPSQ SFPNLFSMAA QRNVTVEEMW DQNFGQGGWN LRFLRAFNDW 180 ELDMVGNLLV EFREHRVTLE EHSVIWKEGG DGLFRVKRAY SVLASGSLFL FDRKVLRYFR 240 KDGHNWRKKK DGKTVKEAHE RLKAGSIDVL HCYYAHGEDN ENFQRRSYWM LEEELSHIVL 300 VHYREVKGNR TSFNRIKETE GALINSQETE EVVPNSETDC SVSSSFPMNS YQMASQTTDT 360 TSLNSAQASE YEDAESAYNH QASSRLHSFL EPVMEKGDAL TAPYYPAPFS NDYQGKLDIP 420 GADFTSLAQE SSSKDSNSVG ISYELPKNLD FPSWEDVLEN CNAGVQSMPS QTPFSSTRAD 480 TMGIIPKQEN EILMQLLTDS FSRKQEFGSD PQGQDEWQTS EGYSAHLSKW PGDQKLHSDS 540 AYGLSTRFDI QEANCVDLLN SLEPGHAYPD GQKANYSSAL KQPLLDSSLT EEGLKKVDSF 600 NRWMSKELGD VNESHMQSRL SSSAAYWDTV ESENGVDESS ISPQGHLDTY MLGPSLSQDQ 660 LFSIIDFSPN WAYAGSEVKV LIMGKFLKGQ QDAEKCKWSC MFGEVEVPAE VISDGVLRCH 720 TPIHKAERVP FYVTCSNRLA CSEVREFEYR VNHIRDVDTA DVSSGSTSEI LLHMRFVKLL 780 SLAPSSNSGL SNEGDRFPLN SKINSLMEED NDEWEQMLML TSEEFSPEKA KEQLLQKLLK 840 EKLHVWLLQK AAEGGKGPNV LDEDGQGVLH FAAALGYDWA IPPTTAAGVS VNFRDVNGWT 900 ALHWAAFCGR ERTVPFLISQ GAAPGALTDP TPKYPAGRTP ADLASSNGHK GIAGYLAESA 960 LSAHLQSLHL KETKEADAAE ISGIKAVQTI SERSPTPIST GDLPLKDSLA AVCNATQAAA 1020 RIHQVFRVQS FQKKQQKEYD DGKFGMSDEH ALSLIAVKSR LGQHDEPVHA AATRIQNKFR 1080 SWKGRKDFLI IRQRIVKIQA HVRGHQVRKN YRKIIWSVGI LEKVILRWRR KGSGLRGFKP 1140 ETHTEGTSMR DISSKEDDYD FLKEGRKQTE ERLQKALARV KSMVQYPEAR DQYRRLLNVV 1200 TEIQETKVVY DRALNSSEEA ADFDDLIDLQ ALLDDDTFMP TAS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4hb5_A | 8e-13 | 853 | 956 | 25 | 122 | Engineered Protein |
| 4hb5_B | 8e-13 | 853 | 956 | 25 | 122 | Engineered Protein |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Vvi.30137 | 0.0 | bud | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in roots, stems, leaves, carpels, and siliques, but not in stigmas or other parts of the flower. {ECO:0000269|PubMed:11162426, ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:14581622}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3'. Binds calmodulin in a calcium-dependent manner in vitro (PubMed:12218065). Regulates transcriptional activity in response to calcium signals (Probable). Involved in freezing tolerance in association with CAMTA1 and CAMTA2 (PubMed:23581962). Required for the cold-induced expression of DREB1B/CBF1, DREB1C/CBF2, ZAT12 and GOLS3 (PubMed:19270186). Involved in response to cold. Contributes together with CAMTA5 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). Involved together with CAMTA2 and CAMTA4 in the positive regulation of a general stress response (GSR) (PubMed:25039701). Involved in the regulation of GSR amplitude downstream of MEKK1 (PubMed:25157030). Involved in the regulation of a set of genes involved in defense responses against pathogens (PubMed:18298954). Involved in the regulation of both basal resistance and systemic acquired resistance (SAR) (PubMed:21900483). Acts as negative regulator of plant immunity (PubMed:19122675, PubMed:21900483, PubMed:22345509, PubMed:28407487). Binds to the promoter of the defense-related gene EDS1 and represses its expression (PubMed:19122675). Binds to the promoter of the defense-related gene NDR1 and represses its expression (PubMed:22345509). Involved in defense against insects (PubMed:23072934, PubMed:22371088). Required for tolerance to the generalist herbivore Trichoplusia ni, and contributes to the positive regulation of genes associated with glucosinolate metabolism (PubMed:23072934). Required for tolerance to Bradysia impatiens larvae. Mediates herbivore-induced wound response (PubMed:22371088). Required for wound-induced jasmonate accumulation (PubMed:23072934, PubMed:22371088). Involved in the regulation of ethylene-induced senescence by binding to the promoter of the senescence-inducer gene EIN3 and repressing its expression (PubMed:22345509). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:18298954, ECO:0000269|PubMed:19122675, ECO:0000269|PubMed:19270186, ECO:0000269|PubMed:21900483, ECO:0000269|PubMed:22345509, ECO:0000269|PubMed:22371088, ECO:0000269|PubMed:23072934, ECO:0000269|PubMed:23581962, ECO:0000269|PubMed:25039701, ECO:0000269|PubMed:25157030, ECO:0000269|PubMed:28351986, ECO:0000269|PubMed:28407487, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00042 | PBM | Transfer from AT2G22300 | Download |
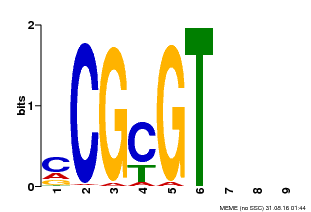
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, salt, wounding, ethylene and methyl jasmonate (PubMed:11162426, PubMed:12218065). Induced by infection with the fungal pathogen Golovinomyces cichoracearum (powdery mildew) and the bacterial pathogen Pseudomonas syringae pv tomato strain DC3000 (PubMed:22345509). {ECO:0000269|PubMed:11162426, ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:22345509}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AM434540 | 0.0 | AM434540.2 Vitis vinifera contig VV78X210461.8, whole genome shotgun sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019073077.1 | 0.0 | PREDICTED: calmodulin-binding transcription activator 3 isoform X2 | ||||
| Swissprot | Q8GSA7 | 0.0 | CMTA3_ARATH; Calmodulin-binding transcription activator 3 | ||||
| TrEMBL | F6HWM2 | 0.0 | F6HWM2_VITVI; Uncharacterized protein | ||||
| STRING | VIT_07s0141g00250.t01 | 0.0 | (Vitis vinifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP562 | 15 | 65 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G22300.2 | 0.0 | signal responsive 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | GSVIVT01004860001 |



