 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSVIVT01008023001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; rosids incertae sedis; Vitales; Vitaceae; Vitis
|
||||||||
| Family | TCP | ||||||||
| Protein Properties | Length: 346aa MW: 38089 Da PI: 8.3349 | ||||||||
| Description | TCP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | TCP | 132.3 | 4.5e-41 | 70 | 175 | 2 | 105 |
TCP 2 agkkdrhskihTkvggRdRRvRlsaecaarfFdLqdeLGfdkdsktieWLlqqakpaikeltgtssssasec.eaesssssasnsssgkaak 92
++k+++++++hTkv+gR+RR+R++a+caar+F+L++eLG+++d++tieWLlqqa+pa++++tgt++++a+++ + s +ss+s+ s +++ +
GSVIVT01008023001 70 PPKRTSTKDRHTKVDGRGRRIRMPALCAARVFQLTRELGHKSDGETIEWLLQQAEPAVIAATGTGTIPANFTsLNISLRSSGSSMSVPSQLR 161
789******************************************************************99988888888888777775555 PP
TCP 93 saa.kskksqksaa 105
s+ ++++s +
GSVIVT01008023001 162 STYfNPNFSVPQRR 175
55444444433332 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03634 | 1.1E-40 | 74 | 209 | IPR005333 | Transcription factor, TCP |
| PROSITE profile | PS51369 | 27.923 | 76 | 130 | IPR017887 | Transcription factor TCP subgroup |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0008283 | Biological Process | cell proliferation | ||||
| GO:0009735 | Biological Process | response to cytokinin | ||||
| GO:0010229 | Biological Process | inflorescence development | ||||
| GO:0010252 | Biological Process | auxin homeostasis | ||||
| GO:0031540 | Biological Process | regulation of anthocyanin biosynthetic process | ||||
| GO:0042023 | Biological Process | DNA endoreduplication | ||||
| GO:0048467 | Biological Process | gynoecium development | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 346 aa Download sequence Send to blast |
MKWDPIDNLS FLRGDELHRP NFPFQLLEKK EEEACSSSGY PSLAISSAEP NTNRSNSSLQ 60 IAAEPSKKPP PKRTSTKDRH TKVDGRGRRI RMPALCAARV FQLTRELGHK SDGETIEWLL 120 QQAEPAVIAA TGTGTIPANF TSLNISLRSS GSSMSVPSQL RSTYFNPNFS VPQRRSLFPG 180 LSSENSSTLT EESIGRKRRP EQDLSQQQMG SYLLQSSTGS IPASHGQIPA NFWMLTNSNN 240 QVMSADPIWP FPSVNNSNLY RGTMSSGLHF MNFPTPVALL PSQQLSSGGG GGSGGGEMKD 300 DIFNMSIDWL VLLNTHLGFY LHTILFPLFV HFFFQELPCN LYALC* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Vvi.26980 | 0.0 | inflorescence | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in archespores, pollen mother cell, ovule primordia and megaspore mother cells (PubMed:25527103). Expressed in young proliferating tissues (PubMed:21668538). Expressed in developing embryos, and seeds during germination (PubMed:25655823). {ECO:0000269|PubMed:21668538, ECO:0000269|PubMed:25527103, ECO:0000269|PubMed:25655823}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved the regulation of plant development. Together with TCP14, modulates plant stature by promoting cell division in young internodes. Represses cell proliferation in developing leaf blade and specific floral tissues (PubMed:21668538). Together with TCP15, acts downstream of gibberellin (GA), and the stratification pathways that promote seed germination. Involved in the control of cell proliferation at the root apical meristem (RAM) by regulating the activity of CYCB1-1 (PubMed:25655823). Acts together with SPY to promote cytokinin responses that affect leaf shape and trichome development in flowers (PubMed:22267487). Involved in gynoecium and silique development. Modulates the development of the different tissues of the gynoecium through its participation in auxin and cytokinin responses. Modulates the expression of the cytokinin-responsive genes ARR7 and ARR15. May repress the expression of the auxin biosynthetic genes YUC1 and YUC4 (PubMed:26303297). Acts as negative regulator of anthocyanin accumulation under high light conditions. Modulates the expression of transcription factors involved in the induction of anthocyanin biosynthesis genes (PubMed:26574599). Transcription factor involved in the regulation of endoreduplication. Represses endoreduplication by activating the gene expression of the key cell-cycle regulators RBR1 and CYCA2-3 (PubMed:25757472). Regulates the expression of the defense gene pathogenesis-related protein 2 (PR2) in antagonism to SRFR1, a negative regulator of effector-triggered immunity (PubMed:24689742). {ECO:0000269|PubMed:21668538, ECO:0000269|PubMed:22267487, ECO:0000269|PubMed:24689742, ECO:0000269|PubMed:25655823, ECO:0000269|PubMed:25757472, ECO:0000269|PubMed:26303297, ECO:0000269|PubMed:26574599}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00224 | DAP | Transfer from AT1G69690 | Download |
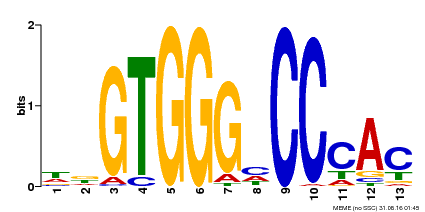
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Circadian-regulation with the lowest expression in the middle of the dark period (PubMed:21183706). Induced during seed imbibition (PubMed:25655823). Induced by cytokinin (PubMed:26303297). {ECO:0000269|PubMed:21183706, ECO:0000269|PubMed:25655823, ECO:0000269|PubMed:26303297}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AM425900 | 0.0 | AM425900.1 Vitis vinifera contig VV78X123770.2, whole genome shotgun sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002268569.1 | 0.0 | PREDICTED: transcription factor TCP15 | ||||
| Swissprot | Q9C9L2 | 1e-66 | TCP15_ARATH; Transcription factor TCP15 | ||||
| TrEMBL | A0A438BV18 | 1e-180 | A0A438BV18_VITVI; Transcription factor TCP15 | ||||
| TrEMBL | F6GT89 | 1e-180 | F6GT89_VITVI; Uncharacterized protein | ||||
| STRING | VIT_17s0000g06020.t01 | 0.0 | (Vitis vinifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP2699 | 12 | 30 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G69690.1 | 4e-50 | TCP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | GSVIVT01008023001 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



