 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSVIVT01009589001 | ||||||||
| Common Name | VIT_18s0001g11390 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; rosids incertae sedis; Vitales; Vitaceae; Vitis
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 288aa MW: 31803.1 Da PI: 8.1731 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 101.4 | 5.9e-32 | 87 | 140 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
pr+rWt++LH++Fv+av+ LGG+e+AtPk++lelm+vk+Ltl+hvkSHLQ+YR+
GSVIVT01009589001 87 PRMRWTTTLHAHFVHAVQLLGGHERATPKSVLELMNVKDLTLAHVKSHLQMYRT 140
9****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.27E-15 | 84 | 141 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 3.1E-28 | 85 | 141 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.7E-23 | 87 | 140 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 1.7E-7 | 88 | 139 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0080060 | Biological Process | integument development | ||||
| GO:0005618 | Cellular Component | cell wall | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 288 aa Download sequence Send to blast |
MFFSSNPIMG TTSPLPDLSL QISPPSISDC EPMLSLGLEM AALNPPPLEL PRNLHHNHHN 60 HQPQIYGREF KRNSRMMNGS KRSIRAPRMR WTTTLHAHFV HAVQLLGGHE RATPKSVLEL 120 MNVKDLTLAH VKSHLQMYRT VKSTDKGTGQ GQTDMGLNQR TGIGQVELGG LSCDKADATP 180 SFSSNTPQPS TPQKISRSSW SLSMETNDEG RLSHENGLKY SHLMANDTKM EGHKVALHVA 240 EGPKERVDSS ALSPSDMLLN LEFTLGRPSW QIDYADQSPN ELALLKC* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 5e-17 | 88 | 142 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 5e-17 | 88 | 142 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 5e-17 | 88 | 142 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 5e-17 | 88 | 142 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 5e-17 | 88 | 142 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 5e-17 | 88 | 142 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 5e-17 | 88 | 142 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 5e-17 | 88 | 142 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00107 | PBM | Transfer from AT5G42630 | Download |
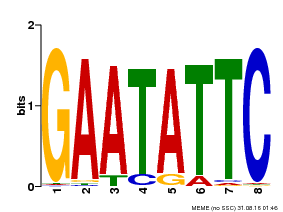
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AM487984 | 1e-133 | AM487984.1 Vitis vinifera, whole genome shotgun sequence, contig VV78X179024.3, clone ENTAV 115. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010664623.1 | 0.0 | PREDICTED: probable transcription factor KAN4 isoform X2 | ||||
| TrEMBL | A0A438K1W0 | 0.0 | A0A438K1W0_VITVI; Putative transcription factor KAN4 | ||||
| STRING | VIT_18s0001g11390.t01 | 0.0 | (Vitis vinifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP570 | 15 | 80 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G42630.1 | 4e-68 | G2-like family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | GSVIVT01009589001 |



