 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSVIVT01010836001 | ||||||||
| Common Name | LOC100266402, VIT_05s0102g01120 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; rosids incertae sedis; Vitales; Vitaceae; Vitis
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 355aa MW: 39316.8 Da PI: 6.2925 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 31.7 | 3.4e-10 | 149 | 207 | 5 | 63 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklksev 63
kr++r NR +A rs +RK +i+eLe+kv++L++e ++L +l l+ l++e+
GSVIVT01010836001 149 KRAKRIWANRQSAARSKERKMRYIAELERKVQTLQTEATSLSAQLTLLQRDTNGLTAEN 207
9*********************************************9998888777776 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 9.8E-16 | 145 | 209 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 10.932 | 147 | 210 | IPR004827 | Basic-leucine zipper domain |
| Gene3D | G3DSA:1.20.5.170 | 1.0E-10 | 149 | 203 | No hit | No description |
| SuperFamily | SSF57959 | 8.44E-11 | 149 | 200 | No hit | No description |
| Pfam | PF00170 | 8.0E-9 | 149 | 203 | IPR004827 | Basic-leucine zipper domain |
| CDD | cd14703 | 4.79E-23 | 150 | 199 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 355 aa Download sequence Send to blast |
MPDNPPKNLG HRRAHSEILT LPDDLSFDSD LGVVGGADGP SFSDETEEDL FSMYLDMDKL 60 NSSSATSSFQ MGEPSSAAPL GSASAAMTEN VVARPNDRRV RHQHSQSMDG STSIKPEMLM 120 SGSDENSAAD SKKAMSAAKL AELALIDPKR AKRIWANRQS AARSKERKMR YIAELERKVQ 180 TLQTEATSLS AQLTLLQRDT NGLTAENSEL KLRLQTMEQQ VNLQDALNDA LKEEIQHLKV 240 LTGQTMPNGA PMMNFPQSYG TGAQFYPNNQ AMHTLLTAQQ FQQLQIHSHK QHQLQQHQLH 300 QLQQQQQQIQ QQIQQQQMQQ EQPQQAGDLK IRGSMASPSQ KENASDTNSS AVKE* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Vvi.4980 | 0.0 | bud| fruit | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Expressed constitutively at a low level in young seedlings and in roots, stems and leaves of mature plants. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Putative transcription factor with an activatory role. | |||||
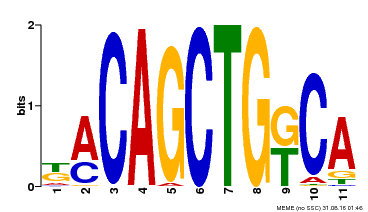
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00126 | DAP | Transfer from AT1G06070 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AM473581 | 0.0 | AM473581.1 Vitis vinifera, whole genome shotgun sequence, contig VV78X277071.8, clone ENTAV 115. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002270784.1 | 0.0 | PREDICTED: probable transcription factor PosF21 | ||||
| Swissprot | Q04088 | 1e-142 | POF21_ARATH; Probable transcription factor PosF21 | ||||
| TrEMBL | F6HYW7 | 0.0 | F6HYW7_VITVI; Uncharacterized protein | ||||
| STRING | VIT_05s0102g01120.t01 | 0.0 | (Vitis vinifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP126 | 17 | 181 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G06070.1 | 1e-130 | bZIP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | GSVIVT01010836001 |
| Entrez Gene | 100266402 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



