 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSVIVT01014236001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; rosids incertae sedis; Vitales; Vitaceae; Vitis
|
||||||||
| Family | TCP | ||||||||
| Protein Properties | Length: 301aa MW: 32904.7 Da PI: 7.5709 | ||||||||
| Description | TCP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | TCP | 115.8 | 5.6e-36 | 42 | 154 | 2 | 89 |
TCP 2 agkkdrhskihTkvggRdRRvRlsaecaarfFdLqdeLGfdkdsktieWLlqqakpaikeltgt................ssssasec.... 73
+g+kdrhsk++T++g+RdRRvRlsa++a++f+d+qd+LG+d++sk+++WL+++ak+ai+el+++ +
GSVIVT01014236001 42 TGRKDRHSKVCTAKGPRDRRVRLSAHTAIQFYDVQDRLGYDRPSKAVDWLIKKAKAAIDELAQLpawhptaitvtaaaaaA------Ttatt 127
79**************************************************************88777777664433220......11111 PP
TCP 74 eaesssssasnsssg.................k 89
+++++ ++ +
GSVIVT01014236001 128 THHQEDEE------NpnqvaidgdddrvqmhhG 154
11111111......0223344555544444321 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03634 | 3.2E-31 | 42 | 147 | IPR005333 | Transcription factor, TCP |
| PROSITE profile | PS51369 | 34.295 | 44 | 102 | IPR017887 | Transcription factor TCP subgroup |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009793 | Biological Process | embryo development ending in seed dormancy | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0030154 | Biological Process | cell differentiation | ||||
| GO:0045962 | Biological Process | positive regulation of development, heterochronic | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 301 aa Download sequence Send to blast |
MAIISIINIL KPNISSSSRM GMRSVVGGEI VEVQGGHIVR STGRKDRHSK VCTAKGPRDR 60 RVRLSAHTAI QFYDVQDRLG YDRPSKAVDW LIKKAKAAID ELAQLPAWHP TAITVTAAAA 120 AATTATTTHH QEDEENPNQV AIDGDDDRVQ MHHGNNPDPN PNPNPNQNPS FLPPSLDSDT 180 IADTIKSFFP MGASGESSSS SVQFQSYPPD LLSRTSSQNQ DLRLSLQSFQ DPMLLHHHHT 240 QSHSHHQQQA LFSPLGFDAP SAAWSEHHAA EMGRHCFAKA RCILRGDPFS PATRLRFALG 300 * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5zkt_A | 8e-21 | 49 | 102 | 1 | 54 | Putative transcription factor PCF6 |
| 5zkt_B | 8e-21 | 49 | 102 | 1 | 54 | Putative transcription factor PCF6 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Vvi.10253 | 0.0 | inflorescence| leaf| root | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Expressed in cotyledons during embryogenesis. Expressed during ovule development (PubMed:25378179). {ECO:0000269|PubMed:25378179}. | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in cotyledons, particularly in the vascular region, in leaves, roots, buds, flowers and immature siliques. {ECO:0000269|PubMed:17307931}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor playing a pivotal role in the control of morphogenesis of shoot organs by negatively regulating the expression of boundary-specific genes such as CUC genes, probably through the induction of miRNA (e.g. miR164) (PubMed:12931144, PubMed:17307931). Required during early steps of embryogenesis (PubMed:15634699). Participates in ovule develpment (PubMed:25378179). Activates LOX2 expression by binding to the 5'-GGACCA-3' motif found in its promoter (PubMed:18816164). {ECO:0000269|PubMed:12931144, ECO:0000269|PubMed:15634699, ECO:0000269|PubMed:17307931, ECO:0000269|PubMed:18816164, ECO:0000269|PubMed:25378179}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00009 | PBM | Transfer from 929286 | Download |
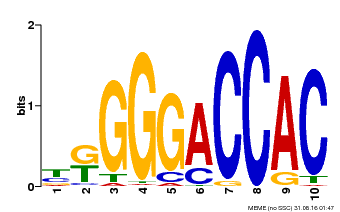
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by the miRNA miR-JAW/miR319 (PubMed:12931144, PubMed:18816164). {ECO:0000269|PubMed:12931144, ECO:0000269|PubMed:18816164}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AM468863 | 0.0 | AM468863.2 Vitis vinifera contig VV78X023076.3, whole genome shotgun sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019072942.1 | 0.0 | PREDICTED: transcription factor TCP4 | ||||
| Swissprot | Q8LPR5 | 1e-78 | TCP4_ARATH; Transcription factor TCP4 | ||||
| TrEMBL | A0A438E5M9 | 0.0 | A0A438E5M9_VITVI; Transcription factor TCP4 | ||||
| TrEMBL | F6H1Y2 | 0.0 | F6H1Y2_VITVI; Uncharacterized protein | ||||
| STRING | VIT_19s0014g01680.t01 | 0.0 | (Vitis vinifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP180 | 15 | 163 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G53230.1 | 8e-50 | TEOSINTE BRANCHED 1, cycloidea and PCF transcription factor 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | GSVIVT01014236001 |



