 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSVIVT01017010001 | ||||||||
| Common Name | VIT_09s0002g03740 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; rosids incertae sedis; Vitales; Vitaceae; Vitis
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 836aa MW: 91996.4 Da PI: 6.4673 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 58.4 | 1.2e-18 | 14 | 72 | 3 | 57 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHC....TS-HHHHHHHHHHHHHHHHC CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkkl....gLterqVkvWFqNrRakekk 57
k ++t+eq+e+Le+l++++++ps +r++L +++ +++ +q+kvWFqNrR +ek+
GSVIVT01017010001 14 KYVRYTPEQVEALERLYHECPKPSSIRRQQLIRECpilsNIEPKQIKVWFQNRRCREKQ 72
5679*****************************************************97 PP
| |||||||
| 2 | START | 181.3 | 5.4e-57 | 158 | 366 | 2 | 205 |
HHHHHHHHHHHHHHHC-TT-EEEEEXCCTTEEEEEEESSS.SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-SEEEEEEEECTT..EEE CS
START 2 laeeaaqelvkkalaeepgWvkssesengdevlqkfeeskvdsgealrasgvvdmvlallveellddkeqWdetlakaetlevissg..gal 91
+aee+++e+++ka+ ++ Wv+++ +++g++++ +++ s++++g a+ra+g+v +++ v+e+l+d++ W ++++++++l+v+ ++ g++
GSVIVT01017010001 158 IAEETLTEFLSKATGTAVEWVQMPGMKPGPDSIGIVAISHGCTGVAARACGLVGLEPT-RVAEILKDRPSWFRDCRAVDVLNVLPTAngGTI 248
789*******************************************************.8888888888****************9999*** PP
EEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--....-TTSEE-EESSEEEEEEEECTCEEEEEEEE-EE--SSXXHH CS
START 92 qlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppe...sssvvRaellpSgiliepksnghskvtwvehvdlkgrlphw 179
+l +++l+a+++l+p Rdf+ +Ry+ +++g++v++++S+ ++q+ p+ +++vRae+lpSg+li+p+++g+s +++v+h+dl+ +++++
GSVIVT01017010001 249 ELLYMQLYAPTTLAPaRDFWLLRYTSVMEDGSLVVCERSLKNTQNGPSmppVQHFVRAEMLPSGYLIRPCEGGGSIIHIVDHMDLEPWSVPE 340
**********************************************9988899*************************************** PP
HHHHHHHHHHHHHHHHHHHHTXXXXX CS
START 180 llrslvksglaegaktwvatlqrqce 205
+lr+l++s+++ ++kt++a+l+++++
GSVIVT01017010001 341 VLRPLYESSTVLAQKTTMAALRQLRQ 366
**********************9876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50071 | 15.418 | 9 | 73 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.2E-15 | 11 | 77 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 5.56E-17 | 14 | 77 | IPR009057 | Homeodomain-like |
| CDD | cd00086 | 9.26E-17 | 14 | 74 | No hit | No description |
| Pfam | PF00046 | 2.9E-16 | 15 | 72 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 1.6E-18 | 16 | 72 | IPR009057 | Homeodomain-like |
| CDD | cd14686 | 1.14E-6 | 66 | 105 | No hit | No description |
| PROSITE profile | PS50848 | 25.584 | 148 | 363 | IPR002913 | START domain |
| CDD | cd08875 | 1.33E-80 | 152 | 368 | No hit | No description |
| SMART | SM00234 | 4.9E-41 | 157 | 367 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 2.88E-38 | 157 | 369 | No hit | No description |
| Gene3D | G3DSA:3.30.530.20 | 3.7E-24 | 157 | 364 | IPR023393 | START-like domain |
| Pfam | PF01852 | 1.7E-54 | 158 | 366 | IPR002913 | START domain |
| Pfam | PF08670 | 5.9E-51 | 692 | 834 | IPR013978 | MEKHLA |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0010014 | Biological Process | meristem initiation | ||||
| GO:0010075 | Biological Process | regulation of meristem growth | ||||
| GO:0010087 | Biological Process | phloem or xylem histogenesis | ||||
| GO:0048263 | Biological Process | determination of dorsal identity | ||||
| GO:0080060 | Biological Process | integument development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 836 aa Download sequence Send to blast |
MSCKDGKGIM DNGKYVRYTP EQVEALERLY HECPKPSSIR RQQLIRECPI LSNIEPKQIK 60 VWFQNRRCRE KQRKEASRLQ AVNRKLTAMN KLLMEENDRL QKQVSQLVYE NGYFRQHTQN 120 TTLATKDTSC ESVVTSGQHH LTPQHPPRDA SPAGLLSIAE ETLTEFLSKA TGTAVEWVQM 180 PGMKPGPDSI GIVAISHGCT GVAARACGLV GLEPTRVAEI LKDRPSWFRD CRAVDVLNVL 240 PTANGGTIEL LYMQLYAPTT LAPARDFWLL RYTSVMEDGS LVVCERSLKN TQNGPSMPPV 300 QHFVRAEMLP SGYLIRPCEG GGSIIHIVDH MDLEPWSVPE VLRPLYESST VLAQKTTMAA 360 LRQLRQIAQE VSQSNVTGWG RRPAALRALS QRLSRGFNEA LNGFTDEGWS MMGNDGIDDV 420 TILVNSSPEK LTGLNLSFAN GFPAVSNAVL CAKASMLLQN VPPAILLRFL REHRSEWADN 480 NIDAYSAAAV KVGPCSLPGS RVGSFGSQVI LPLAHTIEHE EFLEVIKLEG VGHCPEDAMM 540 PRDMFLLQLC SGMDENAVGT CAELIFAPID ASFADDAPLL PSGFRIIPLD SGKEASSPNR 600 TLDLASALEI GPAGNRSSND YSVNGGNTRS VMTIAFEFAF ESHLQENVAS MARQYVRSII 660 SSVQRVALAL SPSHLSSHAG LRPPLGTPEA HTLARWISHS YRCYLGVELL KSSGEGSETI 720 LKTLWHLSDA IMCCSLKALP VFTFANQAGL DMLETTLVAL QDITLEKIFD DHGRKTLCSE 780 FPQIMQQGFA CLQGGICLSS MGRPVSYERA VAWKVLNEEE NAHCVCFMFM NWSFV* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Vvi.16640 | 0.0 | bud | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Highly expressed the developing vascular elements and the adaxial portion of cotyledons. Expressed in developing ovules, stamens and carpels. Expressed in procambium and shoot meristem. {ECO:0000269|PubMed:14701930, ECO:0000269|PubMed:15705957}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the regulation of meristem development to promote lateral organ formation. May regulates procambial and vascular tissue formation or maintenance, and vascular development in inflorescence stems. {ECO:0000269|PubMed:15598805, ECO:0000269|PubMed:15705957, ECO:0000269|PubMed:16617092}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00200 | DAP | Transfer from AT1G52150 | Download |
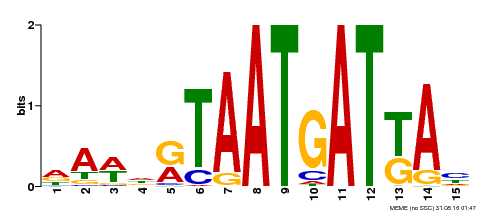
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By auxin. Repressed by miR165 and miR166. {ECO:0000269|PubMed:15773855, ECO:0000269|PubMed:16033795, ECO:0000269|PubMed:16617092, ECO:0000269|PubMed:17237362}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002284003.2 | 0.0 | PREDICTED: homeobox-leucine zipper protein ATHB-15 | ||||
| Swissprot | Q9ZU11 | 0.0 | ATB15_ARATH; Homeobox-leucine zipper protein ATHB-15 | ||||
| TrEMBL | F6HYB4 | 0.0 | F6HYB4_VITVI; Uncharacterized protein | ||||
| STRING | VIT_09s0002g03740.t01 | 0.0 | (Vitis vinifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP651 | 16 | 71 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G52150.1 | 0.0 | HD-ZIP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | GSVIVT01017010001 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



