 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSVIVT01018165001 | ||||||||
| Common Name | LOC100244381, VIT_15s0021g02690 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; rosids incertae sedis; Vitales; Vitaceae; Vitis
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 212aa MW: 23433.4 Da PI: 10.8278 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 50.5 | 3.7e-16 | 33 | 78 | 5 | 55 |
HHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 5 hnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn+ Er+RRd+iN+++ +L++l+P++ K +Ka++L +++eY+k+Lq
GSVIVT01018165001 33 HNQSERKRRDKINQRMKTLQKLVPNS-----SKTDKASMLDEVIEYLKQLQ 78
9************************8.....7******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 6.41E-20 | 25 | 96 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 17.815 | 28 | 77 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 1.4E-18 | 33 | 88 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 3.66E-16 | 33 | 82 | No hit | No description |
| Pfam | PF00010 | 9.6E-14 | 33 | 78 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 2.2E-18 | 34 | 83 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009567 | Biological Process | double fertilization forming a zygote and endosperm | ||||
| GO:0009506 | Cellular Component | plasmodesma | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 212 aa Download sequence Send to blast |
MILKRRAGDE EDKKRGTGKS SVSSKRSRAA AIHNQSERKR RDKINQRMKT LQKLVPNSSK 60 TDKASMLDEV IEYLKQLQAQ VQMMNRMNMS PMMMPMTLQQ QLQMSLMAQM GMGMGMSPMG 120 MGVVDMNTIA RPNVATTGIS PLLHPTPFLP LTSWDVSGDR LPAAPTMVPD PLAAFLACQS 180 QPMTMDAYSR MAALYQHLHQ HPASSARSQG L* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 36 | 41 | ERKRRD |
| 2 | 37 | 42 | RKRRDK |
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in stems and flowers. {ECO:0000269|PubMed:12679534}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Required during the fertilization of ovules by pollen. {ECO:0000269|PubMed:15634699}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00034 | PBM | Transfer from AT4G00050 | Download |
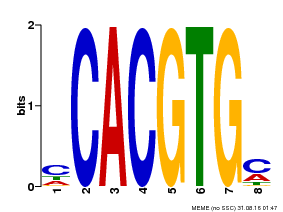
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By cold treatment. {ECO:0000269|PubMed:12679534}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | FQ392071 | 0.0 | FQ392071.1 Vitis vinifera clone SS0AFA7YJ21. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002275629.2 | 1e-147 | PREDICTED: transcription factor UNE10 | ||||
| Swissprot | Q8GZ38 | 3e-59 | UNE10_ARATH; Transcription factor UNE10 | ||||
| TrEMBL | F6GV58 | 1e-146 | F6GV58_VITVI; Uncharacterized protein | ||||
| STRING | VIT_15s0021g02690.t01 | 1e-146 | (Vitis vinifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP8003 | 10 | 14 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00050.1 | 1e-28 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | GSVIVT01018165001 |
| Entrez Gene | 100244381 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



