 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSVIVT01018947001 | ||||||||
| Common Name | VIT_04s0023g01880 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; rosids incertae sedis; Vitales; Vitaceae; Vitis
|
||||||||
| Family | ZF-HD | ||||||||
| Protein Properties | Length: 411aa MW: 46456.5 Da PI: 7.6928 | ||||||||
| Description | ZF-HD family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | ZF-HD_dimer | 105 | 4.8e-33 | 88 | 144 | 3 | 60 |
ZF-HD_dimer 3 kvrYkeClkNhAaslGghavDGCgEfmpsegeegtaaalkCaACgCHRnFHRreveee 60
++rY+eClkNhAas+Ggh++DGCgEfmps geegt +alkCaAC+CHRnFHR+e + e
GSVIVT01018947001 88 SIRYRECLKNHAASMGGHVFDGCGEFMPS-GEEGTLEALKCAACDCHRNFHRKEIDGE 144
689*************************9.999********************98765 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| ProDom | PD125774 | 1.0E-23 | 88 | 142 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| Pfam | PF04770 | 8.6E-31 | 89 | 141 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| TIGRFAMs | TIGR01566 | 4.1E-31 | 90 | 141 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| PROSITE profile | PS51523 | 27.998 | 91 | 140 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| SuperFamily | SSF46689 | 6.16E-18 | 215 | 281 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 3.2E-29 | 218 | 282 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 8.665 | 218 | 282 | IPR001356 | Homeobox domain |
| TIGRFAMs | TIGR01565 | 2.1E-28 | 220 | 277 | IPR006455 | Homeodomain, ZF-HD class |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 411 aa Download sequence Send to blast |
MAEQQAAEIR GRKIMRDIQG VYHRHLHHHQ FNLQQQTQHG EVGDPDPDPD PVSATIAVSG 60 ATATPITGGS NPKVAAAPPH PPPQSAASIR YRECLKNHAA SMGGHVFDGC GEFMPSGEEG 120 TLEALKCAAC DCHRNFHRKE IDGESQPTAN CYYTCNPNTN SSRRNTIAPQ LPPSHAPLPH 180 LHQHHKYSHA PAESSSEDLN MFQSNVGMHL QPQPAFALSK KRFRTKFSQE QKDKMQEFAE 240 KLGWKIQKQE EQEVQQFCSD VGVKRQVFKV WMHNNKQAMK KKQLKTLATS TCIIFLLETY 300 WDLTLSSFHI TSSKSSSPER ERERKEETND KNCTYFPLCL VVMDSWEVSL TYESLFTNLL 360 YYYCTIISCI NPICPFNRQF FGSLKPYHIL LSFSTVEVKQ LDCSCQHTGI * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wh7_A | 1e-21 | 219 | 278 | 16 | 75 | ZF-HD homeobox family protein |
| Search in ModeBase | ||||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in seedlings, roots, leaves, stems, flowers and inflorescence. {ECO:0000269|PubMed:16428600}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Putative transcription factor. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00268 | DAP | Transfer from AT2G18350 | Download |
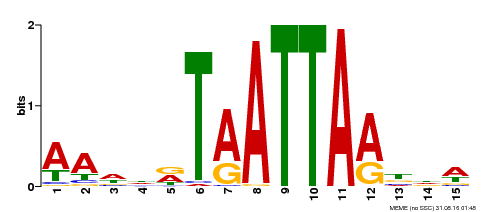
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | FQ384346 | 0.0 | FQ384346.1 Vitis vinifera clone SS0AAG1YN20. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019075027.1 | 0.0 | PREDICTED: zinc-finger homeodomain protein 6 isoform X1 | ||||
| Swissprot | Q9ZPW7 | 1e-62 | ZHD6_ARATH; Zinc-finger homeodomain protein 6 | ||||
| TrEMBL | F6GWV8 | 0.0 | F6GWV8_VITVI; Uncharacterized protein | ||||
| STRING | VIT_04s0023g01880.t01 | 0.0 | (Vitis vinifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP91 | 16 | 237 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G18350.1 | 1e-57 | homeobox protein 24 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | GSVIVT01018947001 |



