 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSVIVT01020814001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; rosids incertae sedis; Vitales; Vitaceae; Vitis
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 545aa MW: 59885 Da PI: 7.4542 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 51.2 | 2.2e-16 | 342 | 388 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K +Ka++L +A+eY+ksLq
GSVIVT01020814001 342 VHNLSERRRRDRINEKMKALQELIPHS-----NKSDKASMLDEAIEYLKSLQ 388
6*************************7.....59*****************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 1.23E-19 | 336 | 398 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 17.942 | 338 | 387 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 1.16E-14 | 341 | 386 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 3.1E-19 | 342 | 395 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 8.2E-14 | 342 | 388 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 3.4E-17 | 344 | 393 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009585 | Biological Process | red, far-red light phototransduction | ||||
| GO:0009693 | Biological Process | ethylene biosynthetic process | ||||
| GO:0010244 | Biological Process | response to low fluence blue light stimulus by blue low-fluence system | ||||
| GO:0010600 | Biological Process | regulation of auxin biosynthetic process | ||||
| GO:0010928 | Biological Process | regulation of auxin mediated signaling pathway | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 545 aa Download sequence Send to blast |
MNHPLLPDWN IEGDVPVTNQ KKPMGQEHEL VELLWRNGQV VLHSQTQRKS GLAPNESKQV 60 QKHDQTMLRN GGSCGNWSNL IQDEESISWI QYPLDDSLDK EFCSNLFMEL PPNDPVQPEK 120 PVRHSEQEKS VPSSQQPIFK HLGSPEFPGN PMPPPKFQVP GSVQQNCSSG GFGKIVNFPN 180 FSFPGRSNLG SSKAQLGGKG SGNMAQGGDA KEGSVMTVGL SHCGSNQVVN EADLSRFSSS 240 GVGTGCLSSG HVKENVMRIV SQGDRRQTET LEPTVTSSSG GGSGSSFGRT YKQSTDTNSH 300 KRKGREAEES ECQSEAAEHE SAARNKASQR SGSTRRSRAA EVHNLSERRR RDRINEKMKA 360 LQELIPHSNK SDKASMLDEA IEYLKSLQLQ LQLMWMGGGV APMMFPGVQH YMARMGMGMC 420 PPPLPSIHNP MHLPRVQLVD QSTSAVPPSN QPPICQTPVL NPVNYQNQMP NPNFPEQFAH 480 YMGFHPMQTP SQPMNVFSFG SQTSWVNTSG YACCSNAFGS ITHQVRPLQD DDKSHPLISS 540 WWMA* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 346 | 351 | ERRRRD |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Vvi.16948 | 0.0 | inflorescence| leaf | ||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00082 | ChIP-seq | Transfer from AT3G59060 | Download |
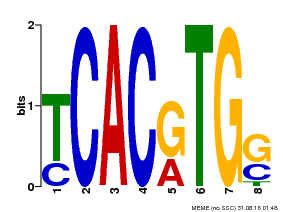
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AM468168 | 0.0 | AM468168.2 Vitis vinifera contig VV78X251080.4, whole genome shotgun sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002278399.1 | 0.0 | PREDICTED: transcription factor PIF4 | ||||
| Refseq | XP_010657098.1 | 0.0 | PREDICTED: transcription factor PIF4 | ||||
| TrEMBL | F6H570 | 0.0 | F6H570_VITVI; Uncharacterized protein | ||||
| STRING | VIT_12s0028g01110.t01 | 0.0 | (Vitis vinifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP258 | 16 | 128 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G59060.1 | 3e-54 | phytochrome interacting factor 3-like 6 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | GSVIVT01020814001 |



