 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSVIVT01021098001 | ||||||||
| Common Name | VIT_10s0003g00140 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; rosids incertae sedis; Vitales; Vitaceae; Vitis
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 228aa MW: 24589.4 Da PI: 10.03 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 64 | 3.1e-20 | 20 | 70 | 2 | 56 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkklege 56
+++GVr ++ +gr++AeIrdp gk+ r++lg+f+taeeAaka++ a+++++g+
GSVIVT01021098001 20 HFRGVRKRP-WGRYAAEIRDP---GKKSRVWLGTFDTAEEAAKAYDSAAREFRGA 70
69*******.**********8...557*************************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF54171 | 1.05E-21 | 20 | 78 | IPR016177 | DNA-binding domain |
| PROSITE profile | PS51032 | 23.525 | 20 | 77 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 3.9E-32 | 20 | 78 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 3.08E-30 | 20 | 77 | No hit | No description |
| SMART | SM00380 | 1.3E-38 | 20 | 83 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 1.1E-13 | 20 | 69 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 7.7E-11 | 21 | 32 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 7.7E-11 | 43 | 59 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009864 | Biological Process | induced systemic resistance, jasmonic acid mediated signaling pathway | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0016604 | Cellular Component | nuclear body | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 228 aa Download sequence Send to blast |
MAPKEKVAGV KPSANAKEVH FRGVRKRPWG RYAAEIRDPG KKSRVWLGTF DTAEEAAKAY 60 DSAAREFRGA KAKTNFPLVS ENLNNNNQSP SQSSTVESSS REGFSPALMV DSSPLDLNLL 120 HGGGVGHHQF QVSSPSPAAG IVPTGGLPAA NHLFYFDAML RTGRVNQDFQ RLRFDRAASD 180 FRAALTGGVQ SDSDSSSVVD LNHNDLKPRA RVLIDLDLNR PPPPEIA* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2gcc_A | 4e-22 | 16 | 78 | 1 | 64 | ATERF1 |
| 3gcc_A | 4e-22 | 16 | 78 | 1 | 64 | ATERF1 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Vvi.581 | 0.0 | bud| flower| fruit| inflorescence| pedicel| root | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in roots, mostly in root tip, lateral root tips, cortex and vascular tissues. {ECO:0000269|PubMed:10945353}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds to the GCC-box pathogenesis-related promoter element. Involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways. Probably acts as a transcriptional repressor and may regulate other AtERFs (By similarity). {ECO:0000250}. | |||||
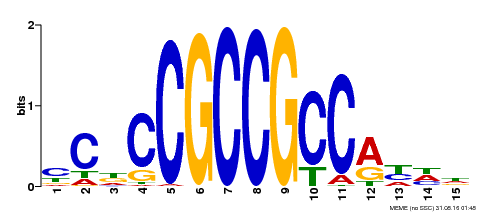
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00358 | DAP | Transfer from AT3G15210 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Strongly and quickly induced by ethylene. {ECO:0000269|PubMed:10945353}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY395744 | 0.0 | AY395744.1 Vitis aestivalis putative ethylene response factor ERF3a mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002279692.1 | 1e-137 | PREDICTED: ethylene-responsive transcription factor 4 | ||||
| Swissprot | Q9LW49 | 2e-45 | ERF4_NICSY; Ethylene-responsive transcription factor 4 | ||||
| TrEMBL | D7TJC7 | 1e-164 | D7TJC7_VITVI; Uncharacterized protein | ||||
| STRING | VIT_10s0003g00140.t01 | 1e-165 | (Vitis vinifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP6 | 16 | 1718 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G44210.1 | 2e-27 | erf domain protein 9 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | GSVIVT01021098001 |



