 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSVIVT01023585001 | ||||||||
| Common Name | VIT_11s0037g00040 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; rosids incertae sedis; Vitales; Vitaceae; Vitis
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 283aa MW: 29322.7 Da PI: 7.0435 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 37.9 | 3.2e-12 | 115 | 160 | 5 | 55 |
HHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 5 hnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
h+++Er RR+ri +++ L+el+P+a K +Ka++L + ++Y+k Lq
GSVIVT01023585001 115 HSIAERLRRERIAERMKALQELVPNA-----NKTDKASMLDEIIDYVKFLQ 160
99***********************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 2.49E-17 | 108 | 170 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 6.59E-14 | 108 | 164 | No hit | No description |
| PROSITE profile | PS50888 | 17.39 | 110 | 159 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 8.6E-17 | 112 | 169 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.5E-9 | 115 | 160 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.5E-13 | 116 | 165 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045944 | Biological Process | positive regulation of transcription from RNA polymerase II promoter | ||||
| GO:0048767 | Biological Process | root hair elongation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0001046 | Molecular Function | core promoter sequence-specific DNA binding | ||||
| GO:0001228 | Molecular Function | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 283 aa Download sequence Send to blast |
MLQQQLLFNG DSCLVDRSQN DVVDGSSSFK SPNQGGDGSV QALYNGFAGA LHGSGQASNQ 60 AQNFHHPQGG SMQAQNYGAP ATVMNQTPAT GSAGGAPAQP RQRVRARRGQ ATDPHSIAER 120 LRRERIAERM KALQELVPNA NKTDKASMLD EIIDYVKFLQ LQVKVLSMSR LGGAAAVAPL 180 VADMSSEGGG DCIQASGTSG PTGGRATNGT QTTTSNDSLT VTEHQVAKLM EEDMGSAMQY 240 LQGKGLCLMP ISLATAISTT TSTMGNGLAD APVKDAASVS KP* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 119 | 124 | RLRRER |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Vvi.18419 | 0.0 | fruit| leaf| root | ||||
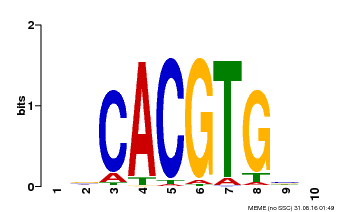
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00659 | PBM | Transfer from Pp3c14_15890 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By cold, UV, ethylene (ACC), flagellin, jasmonic acid (JA), and salicylic acid (SA) treatments. {ECO:0000269|PubMed:12679534}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AM486521 | 1e-118 | AM486521.2 Vitis vinifera contig VV78X151465.9, whole genome shotgun sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010656596.1 | 0.0 | PREDICTED: transcription factor MYC2 isoform X1 | ||||
| Swissprot | Q9ZUG9 | 1e-84 | BH066_ARATH; Transcription factor bHLH66 | ||||
| TrEMBL | F6HYI3 | 0.0 | F6HYI3_VITVI; Uncharacterized protein | ||||
| STRING | VIT_11s0037g00040.t01 | 0.0 | (Vitis vinifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP58 | 16 | 313 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G24260.1 | 2e-47 | LJRHL1-like 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | GSVIVT01023585001 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



