 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSVIVT01025324001 | ||||||||
| Common Name | LOC100248434, VIT_06s0004g01610 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; rosids incertae sedis; Vitales; Vitaceae; Vitis
|
||||||||
| Family | EIL | ||||||||
| Protein Properties | Length: 590aa MW: 66669.7 Da PI: 6.3918 | ||||||||
| Description | EIL family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | EIN3 | 427.6 | 1.2e-130 | 38 | 377 | 1 | 342 |
XXXXXXXXXXXXXXXXXXXXXXX..XXXXX.XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX CS
EIN3 1 eelkkrmwkdqmllkrlkerkkqlledkeaatgakksnksneqarrkkmsraQDgiLkYMlkemevcnaqGfvYgiipekgkpvegasdsLr 92
eel++rmwkd+++lkr+ker+k +++a++++k + ++arrkkmsraQDgiLkYMlk mevc+a+GfvYgiipekgkpv+gasd++r
GSVIVT01025324001 38 EELERRMWKDRIKLKRIKERQKIT---AQQAAEKQKPKPNADHARRKKMSRAQDGILKYMLKLMEVCKARGFVYGIIPEKGKPVSGASDNIR 126
8*****************999964...34458999********************************************************* PP
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX....XX----STTS-HHHHHHHHHHHSSSSSS-TTS--TTT--HHHH---S--HHHHHH CS
EIN3 93 aWWkekvefdrngpaaiskyqaknlilsgesslqtersseshslselqDTtlgSLLsalmqhcdppqrrfplekgvepPWWPtGkelwwgel 184
aWWkekv+fd+ngpaai+ky+a++l++ +++++ ++ +++++l++lqD+tlgSLLs+lmqhcdppqr++plekgv+pPWWP+G+e+ww +l
GSVIVT01025324001 127 AWWKEKVKFDKNGPAAIAKYEAECLAMVENENN--RNGNSQSTLQDLQDATLGSLLSSLMQHCDPPQRKYPLEKGVPPPWWPSGNEDWWVKL 216
**************************9988888..689999*************************************************** PP
T--TT--.-----GGG--HHHHHHHHHHHHHHTGGGHHHHHHTTTTSSSSTTT--SHHHHHHHHHHTTTTT-S--XXXXXX.........XX CS
EIN3 185 glskdqgtppykkphdlkkawkvsvLtavikhmsptieeirelerqskylqdkmsakesfallsvlnqeekecatvsahss.........sl 267
gl+++q+ ppykkphdlkk+wkv+vLtavikhmsp+i++ir+l+rqsk+lqdkm+akes ++l vln+ee+++++ s+++ +
GSVIVT01025324001 217 GLARSQS-PPYKKPHDLKKMWKVGVLTAVIKHMSPDISKIRRLVRQSKCLQDKMTAKESSIWLGVLNREESLIRQPSSDNGtsgitgtppNG 307
******9.9**********************************************************************5543455544433 PP
XXXXXXXXXXXXXXXXXXXXXXXX.XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX......XXXXXXX.XXXXX CS
EIN3 268 rkqspkvtlsceqkedvegkkeskikhvqavkttagfpvvrkrkkkpsesakvsskevsrtcqssqfrgseteli 342
+++++kv ++ + e +++ + +q v+ +++ + krk++ ++s+ v+++++ ++ ++ + ++ ++
GSVIVT01025324001 308 HDGKNKVAMDIASFE-----DDRDNSGAQPVQDKKKGRKQLKRKRPHVKSRPVNQESAPSLEENLHEESRNSLQH 377
334444444433332.....2222244444444445556666666666666666665544444444433333222 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04873 | 5.1E-127 | 38 | 285 | No hit | No description |
| Gene3D | G3DSA:1.10.3180.10 | 3.2E-72 | 159 | 293 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| SuperFamily | SSF116768 | 3.4E-62 | 166 | 289 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009873 | Biological Process | ethylene-activated signaling pathway | ||||
| GO:0042762 | Biological Process | regulation of sulfur metabolic process | ||||
| GO:0071281 | Biological Process | cellular response to iron ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 590 aa Download sequence Send to blast |
MGEFEEIGVD MCSDIEVDEV RCENIAEKDV SDEEIEAEEL ERRMWKDRIK LKRIKERQKI 60 TAQQAAEKQK PKPNADHARR KKMSRAQDGI LKYMLKLMEV CKARGFVYGI IPEKGKPVSG 120 ASDNIRAWWK EKVKFDKNGP AAIAKYEAEC LAMVENENNR NGNSQSTLQD LQDATLGSLL 180 SSLMQHCDPP QRKYPLEKGV PPPWWPSGNE DWWVKLGLAR SQSPPYKKPH DLKKMWKVGV 240 LTAVIKHMSP DISKIRRLVR QSKCLQDKMT AKESSIWLGV LNREESLIRQ PSSDNGTSGI 300 TGTPPNGHDG KNKVAMDIAS FEDDRDNSGA QPVQDKKKGR KQLKRKRPHV KSRPVNQESA 360 PSLEENLHEE SRNSLQHLHN EPRNSLPDIN HNDAQLAPYE MLGTQQENDR VTSLRPLEKD 420 LENQSQLPEP EFNHFSAFPS ANAISTQSMY VGGRPLLYPA VQNAELHHGT PYEFYNPPSD 480 YGHNPDGQQS HMAMNETQMR LEDGRIHEPE LNRNGNDISG GNLRHYVKDT FHSGQDRPVE 540 SHYGSPIESL SLDFGGFNSP FDLGIDGTSL GTPDIDYLLD DDLIQYFGA* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wij_A | 2e-74 | 165 | 296 | 9 | 140 | ETHYLENE-INSENSITIVE3-like 3 protein |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Vvi.22412 | 0.0 | cell culture | ||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may be involved in the ethylene response pathway. {ECO:0000269|PubMed:9215635, ECO:0000269|PubMed:9851977}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00233 | DAP | Transfer from AT1G73730 | Download |
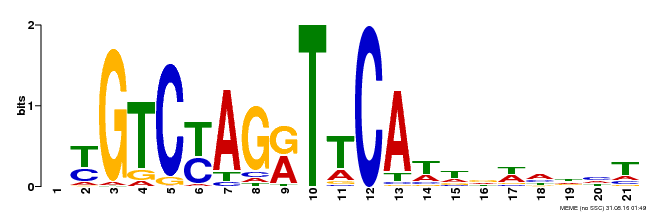
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AM471537 | 0.0 | AM471537.1 Vitis vinifera, whole genome shotgun sequence, contig VV78X037171.18, clone ENTAV 115. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019076322.1 | 0.0 | PREDICTED: ETHYLENE INSENSITIVE 3-like 3 protein isoform X2 | ||||
| Swissprot | O23116 | 0.0 | EIL3_ARATH; ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| TrEMBL | F6GU99 | 0.0 | F6GU99_VITVI; Uncharacterized protein | ||||
| STRING | VIT_06s0004g01610.t01 | 0.0 | (Vitis vinifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP563 | 16 | 82 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G73730.1 | 0.0 | ETHYLENE-INSENSITIVE3-like 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | GSVIVT01025324001 |
| Entrez Gene | 100248434 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



