 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSVIVT01027508001 | ||||||||
| Common Name | LOC100260889, VIT_15s0048g02000 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; rosids incertae sedis; Vitales; Vitaceae; Vitis
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 772aa MW: 83910.4 Da PI: 6.1814 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 65.7 | 6.3e-21 | 113 | 168 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ +++t++q++eLe+lF+++++p++++r eL+++l+L++rqVk+WFqNrR+++k
GSVIVT01027508001 113 KKRYHRHTPQQIQELEALFKECPHPDEKQRLELSRRLSLETRQVKFWFQNRRTQMK 168
688999***********************************************999 PP
| |||||||
| 2 | START | 208.1 | 3.4e-65 | 281 | 505 | 1 | 206 |
HHHHHHHHHHHHHHHHC-TT-EEEE....EXCCTTEEEEEEESSS......SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S....EE CS
START 1 elaeeaaqelvkkalaeepgWvkss....esengdevlqkfeeskv.....dsgealrasgvvdmvlallveellddkeqWdetla....ka 79
ela++a++elvk+a+ +ep+Wv+s e++n +e++++f++ + + +e r++g+v+ ++ lve+l+d++ +W e+++ +
GSVIVT01027508001 281 ELALAAMDELVKMAQTDEPLWVRSLeggrEILNLEEYMRTFTPCIGmkpsgFVTESTRETGMVIINSLALVETLMDSN-RWAEMFPcmiaRT 371
5899**************************************9998999999**************************.************* PP
EEEEEECTT......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--.-TTSEE-EESSEEEEEEEECTCEEEE CS
START 80 etlevissg......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppesssvvRaellpSgiliepksnghskv 164
+t++vissg galqlm aelq+lsplvp R++ f+R+++q+ +g+w++vdvS+d ++ + + +v +++lpSg+++++++ng+skv
GSVIVT01027508001 372 STTDVISSGmggtrnGALQLMHAELQVLSPLVPvREVNFLRFCKQHAEGVWAVVDVSIDTIRETSVAPTFVNCRRLPSGCVVQDMPNGYSKV 463
***************************************************************999************************** PP
EEEE-EE--SSXXHHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 165 twvehvdlkgrlphwllrslvksglaegaktwvatlqrqcek 206
twveh++++++ +h+l+r+l++sg+ +ga++wvatlqrqce+
GSVIVT01027508001 464 TWVEHAEYDESAVHQLYRPLLGSGMGFGAQRWVATLQRQCEC 505
****************************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 8.77E-21 | 96 | 170 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 7.6E-22 | 99 | 164 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.605 | 110 | 170 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 4.1E-19 | 111 | 174 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 5.20E-19 | 112 | 170 | No hit | No description |
| Pfam | PF00046 | 1.7E-18 | 113 | 168 | IPR001356 | Homeobox domain |
| PROSITE pattern | PS00027 | 0 | 145 | 168 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 43.25 | 272 | 508 | IPR002913 | START domain |
| CDD | cd08875 | 1.03E-127 | 277 | 504 | No hit | No description |
| SuperFamily | SSF55961 | 2.38E-34 | 277 | 505 | No hit | No description |
| SMART | SM00234 | 9.6E-50 | 281 | 505 | IPR002913 | START domain |
| Pfam | PF01852 | 8.6E-57 | 281 | 505 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 5.63E-24 | 534 | 764 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009827 | Biological Process | plant-type cell wall modification | ||||
| GO:0042335 | Biological Process | cuticle development | ||||
| GO:0043481 | Biological Process | anthocyanin accumulation in tissues in response to UV light | ||||
| GO:0048765 | Biological Process | root hair cell differentiation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 772 aa Download sequence Send to blast |
MSFGGFLDNS SGGGGARIVA DIPYSNNMAT GAIAQPRLVS PSLAKSMFSS PGLSLALQTS 60 MEGQGEVTRL AENFESGGGR RSREDEHESR SGSDNMDGAS GDDQDAADNP PRKKRYHRHT 120 PQQIQELEAL FKECPHPDEK QRLELSRRLS LETRQVKFWF QNRRTQMKTQ LERHENSILR 180 QENDKLRAEN MSIRDAMRNP ICTNCGGPAI IGDISLEEQH LRIENARLKD ELDRVCALAG 240 KFLGRPISSL ASSMAPAMPS SSLELGVGSN GGISSTSMFL ELALAAMDEL VKMAQTDEPL 300 WVRSLEGGRE ILNLEEYMRT FTPCIGMKPS GFVTESTRET GMVIINSLAL VETLMDSNRW 360 AEMFPCMIAR TSTTDVISSG MGGTRNGALQ LMHAELQVLS PLVPVREVNF LRFCKQHAEG 420 VWAVVDVSID TIRETSVAPT FVNCRRLPSG CVVQDMPNGY SKVTWVEHAE YDESAVHQLY 480 RPLLGSGMGF GAQRWVATLQ RQCECLAILM SSTVPTRDHT AAITAGGRRS MLKLAQRMTD 540 NFCAGVCAST VHKWNKLCAG NVDEDVRVMT RKSVDDPGEP PGIVLSAATS VWLPVSPQRL 600 FDFLRDERLR SEWDILSNGG PMQEMAHIAK GQDHGNCVSL LRASAMNANQ SSMLILQETC 660 IDAAGSLVVY APVDIPAMHV VMNGGDSAYV ALLPSGFAIV PDGPGSRGPN SGVHTNSGGP 720 NRVSGSLLTV AFQILVNSLP TAKLTVESVE TVNNLISCTV QKIKAALHCE S* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Vvi.11959 | 0.0 | bud| flower| fruit | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in roots, stems, leaves and floral buds. {ECO:0000269|PubMed:10402424}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the regulation of the tissue-specific accumulation of anthocyanins and in cellular organization of the primary root. {ECO:0000269|PubMed:10402424}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00421 | DAP | Transfer from AT4G00730 | Download |
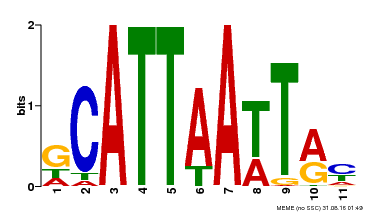
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AM459307 | 0.0 | AM459307.2 Vitis vinifera contig VV78X101111.6, whole genome shotgun sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002272264.2 | 0.0 | PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 isoform X1 | ||||
| Refseq | XP_010661561.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 isoform X1 | ||||
| Swissprot | Q0WV12 | 0.0 | ANL2_ARATH; Homeobox-leucine zipper protein ANTHOCYANINLESS 2 | ||||
| TrEMBL | F6I316 | 0.0 | F6I316_VITVI; Uncharacterized protein | ||||
| STRING | VIT_15s0048g02000.t01 | 0.0 | (Vitis vinifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP145 | 15 | 136 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00730.1 | 0.0 | HD-ZIP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | GSVIVT01027508001 |
| Entrez Gene | 100260889 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



