 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSVIVT01031089001 | ||||||||
| Common Name | VIT_14s0060g02660 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; rosids incertae sedis; Vitales; Vitaceae; Vitis
|
||||||||
| Family | NF-YB | ||||||||
| Protein Properties | Length: 148aa MW: 16544.8 Da PI: 7.3415 | ||||||||
| Description | NF-YB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NF-YB | 162.6 | 5.7e-51 | 3 | 98 | 2 | 97 |
NF-YB 2 reqdrflPianvsrimkkvlPanakiskdaketvqecvsefisfvtseasdkcqrekrktingddllwalatlGfedyveplkvylkkyreleg 95
eqd++lPianv+rimk++lP +akisk+ ket+qec+sefisfvt+easdkc++e+rkt+ngdd++wal+ lGf+dy+e++ yl+kyre+e+
GSVIVT01031089001 3 DEQDHLLPIANVGRIMKQILPPRAKISKEGKETMQECASEFISFVTGEASDKCHKENRKTVNGDDICWALSALGFDDYAEAILRYLHKYREFER 96
69*******************************************************************************************9 PP
NF-YB 96 ek 97
e+
GSVIVT01031089001 97 ER 98
97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.20.10 | 3.4E-48 | 2 | 112 | IPR009072 | Histone-fold |
| SuperFamily | SSF47113 | 1.31E-37 | 5 | 113 | IPR009072 | Histone-fold |
| Pfam | PF00808 | 2.3E-25 | 9 | 72 | IPR003958 | Transcription factor CBF/NF-Y/archaeal histone domain |
| PRINTS | PR00615 | 7.2E-18 | 36 | 54 | No hit | No description |
| PROSITE pattern | PS00685 | 0 | 39 | 55 | IPR003956 | Transcription factor, NFYB/HAP3, conserved site |
| PRINTS | PR00615 | 7.2E-18 | 55 | 73 | No hit | No description |
| PRINTS | PR00615 | 7.2E-18 | 74 | 92 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0046982 | Molecular Function | protein heterodimerization activity | ||||
| Plant Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PO Term | PO Category | PO Description | ||||
| PO:0004709 | anatomy | axillary bud | ||||
| PO:0025532 | developmental stage | bud burst stage | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 148 aa Download sequence Send to blast |
MVDEQDHLLP IANVGRIMKQ ILPPRAKISK EGKETMQECA SEFISFVTGE ASDKCHKENR 60 KTVNGDDICW ALSALGFDDY AEAILRYLHK YREFERERAN QNKVGGSEDK DEASNCKYIQ 120 AGKQTVIPPP LECRVIKKGN SSLTKPS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4g91_B | 5e-40 | 2 | 93 | 1 | 92 | Transcription factor HapC (Eurofung) |
| 4g92_B | 5e-40 | 2 | 93 | 1 | 92 | Transcription factor HapC (Eurofung) |
| Search in ModeBase | ||||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in flowers and siliques. {ECO:0000269|PubMed:11250072}. | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in flowers, siliques and young rosettes. {ECO:0000269|PubMed:11250072}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Component of the NF-Y/HAP transcription factor complex. The NF-Y complex stimulates the transcription of various genes by recognizing and binding to a CCAAT motif in promoters. | |||||
| UniProt | Component of the NF-Y/HAP transcription factor complex. The NF-Y complex stimulates the transcription of various genes by recognizing and binding to a CCAAT motif in promoters. | |||||
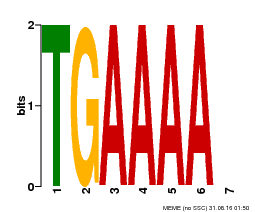
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00133 | DAP | Transfer from AT1G09030 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AM480950 | 0.0 | AM480950.1 Vitis vinifera, whole genome shotgun sequence, contig VV78X208183.15, clone ENTAV 115. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019080620.1 | 1e-108 | PREDICTED: nuclear transcription factor Y subunit B-4 | ||||
| Swissprot | O04027 | 2e-54 | NFYB4_ARATH; Nuclear transcription factor Y subunit B-4 | ||||
| Swissprot | O82248 | 3e-54 | NFYB5_ARATH; Nuclear transcription factor Y subunit B-5 | ||||
| TrEMBL | D7U9R6 | 1e-106 | D7U9R6_VITVI; Uncharacterized protein | ||||
| STRING | VIT_14s0060g02660.t01 | 1e-107 | (Vitis vinifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP168 | 17 | 170 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09030.1 | 8e-57 | nuclear factor Y, subunit B4 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | GSVIVT01031089001 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



