 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSVIVT01035027001 | ||||||||
| Common Name | LOC100243111, VIT_05s0077g01240 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; rosids incertae sedis; Vitales; Vitaceae; Vitis
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 928aa MW: 104866 Da PI: 7.3047 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 160.2 | 4e-50 | 30 | 146 | 3 | 118 |
CG-1 3 ke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenpt 93
+e k rwl+++ei+aiL n++ +++ ++ + p+sg+++L++r+++r+frkDG++wkkk dgktv+E+he+LKvg+ e +++yYah+++npt
GSVIVT01035027001 30 EEaKGRWLRPNEIHAILCNYTLFTVNVKPVNLPPSGKIVLFDRRMLRNFRKDGHNWKKKNDGKTVKEAHEHLKVGNDERIHVYYAHGQDNPT 121
45599*************************************************************************************** PP
CG-1 94 fqrrcywlLeeelekivlvhylevk 118
f rrcywlL+++le+ivlvhy+e++
GSVIVT01035027001 122 FVRRCYWLLDKTLEHIVLVHYRETQ 146
**********************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 77.632 | 25 | 151 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 1.5E-73 | 28 | 146 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 3.9E-45 | 31 | 144 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 6.3E-4 | 371 | 471 | IPR013783 | Immunoglobulin-like fold |
| SuperFamily | SSF81296 | 4.06E-14 | 383 | 469 | IPR014756 | Immunoglobulin E-set |
| Pfam | PF12796 | 3.8E-8 | 562 | 647 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 1.03E-17 | 569 | 677 | No hit | No description |
| SuperFamily | SSF48403 | 8.24E-18 | 569 | 684 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 6.6E-19 | 575 | 682 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 18.431 | 576 | 681 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 2.6E-5 | 618 | 647 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 11.594 | 618 | 650 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 240 | 657 | 688 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 3.98E-6 | 763 | 832 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| PROSITE profile | PS50096 | 7.254 | 766 | 792 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 30 | 781 | 803 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.785 | 782 | 811 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0026 | 804 | 826 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 10.237 | 805 | 829 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 6.9E-4 | 806 | 826 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 9.5 | 880 | 902 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.206 | 882 | 910 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045944 | Biological Process | positive regulation of transcription from RNA polymerase II promoter | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0001077 | Molecular Function | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 928 aa Download sequence Send to blast |
MESSVPGRLA GWDIHGFRTM EDLDVDSILE EAKGRWLRPN EIHAILCNYT LFTVNVKPVN 60 LPPSGKIVLF DRRMLRNFRK DGHNWKKKND GKTVKEAHEH LKVGNDERIH VYYAHGQDNP 120 TFVRRCYWLL DKTLEHIVLV HYRETQESQG SPVTPVNSSP SPNSATSDPS APWLLSEETD 180 SGTGSTYRAG EKEHQEPRDS ITVRNYEMRI HELNTLEWDE LLVSNDPNNS MAPKEGKISS 240 FEQQNQHVIT SSNSYNRPHS TNDLPVGISP LGNPAESIAG NESAHFNFLD DVYFQKIGGQ 300 VNPNGQRRDS VAVGTGDPVD ILLKDSLEPQ DSFGRWMNYI MTDSPVSVDD PSLGSPVSSS 360 HDSVVSAAGN HQQSSVPDTI FSITDFSPSW AISTEKTKIL VIGFLHENYA DLAKSNLFFV 420 CGDVCVPAEI IQLGVFRCLV PPHAPGLVNF YLSFDGHKPI SQVVTFEYRA PLLYNQTVSS 480 EVETNWEEFQ FQMRLSHLLF STSKGLNIMS SKISPNALRE AKNFVKKTSF IARNWANLTK 540 TIGDNRILVS QAKDLLFEFA LLNKLQEWLV ERIVEGGKTS ERDGQGQGVI HLCAMLGYTR 600 AVYLYSLSGL SLDYRDKFGW TALHWAAYYG RQKMVAVLLS AGAKPNLVTD PTSENPGGCT 660 AADLASKEGH DGLAAYLAEK GLVEQFNDMT LAGNVSGSLQ VSTTEQINSE NLSEEEMNLK 720 DTLAAYRTAA DAAARIQVAF RERSLKLRTK AVENCNPEIE ARNIVAAMRI QHAFRNYETR 780 KRMAAAARIQ HRFRSWKIRK EFLNMRRQAI KIQAVFRGFQ VRRQYRKILW SVGVLEKVIL 840 RWRMKRKGFR GLQVDTVDQL QESDTEEDFF RASRRQAEDR VERSVIRVQA MFRSKKAQEE 900 YRRMKLAHNE AKLEFEGFID PDTNMDG* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Vvi.10386 | 0.0 | bud| flower| fruit| inflorescence| leaf | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in roots, stems, leaves, pollen, top of sepals and siliques. {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:14581622}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator (PubMed:14581622). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in response to cold. Contributes together with CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:28351986, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
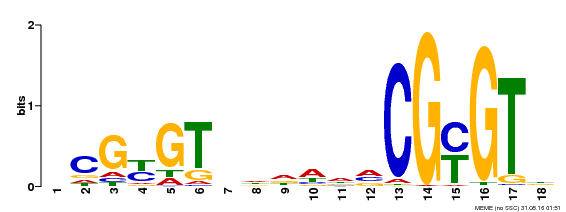
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, wounding, abscisic acid, H(2)O(2) and salicylic acid. {ECO:0000269|PubMed:12218065}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | FQ381725 | 0.0 | FQ381725.1 Vitis vinifera clone SS0ABG86YO23. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002272118.2 | 0.0 | PREDICTED: calmodulin-binding transcription activator 6 isoform X1 | ||||
| Swissprot | O23463 | 0.0 | CMTA5_ARATH; Calmodulin-binding transcription activator 5 | ||||
| TrEMBL | D7SY49 | 0.0 | D7SY49_VITVI; Uncharacterized protein | ||||
| STRING | VIT_05s0077g01240.t01 | 0.0 | (Vitis vinifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP7351 | 11 | 16 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | GSVIVT01035027001 |
| Entrez Gene | 100243111 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



