 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | GSVIVT01036911001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; rosids incertae sedis; Vitales; Vitaceae; Vitis
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 613aa MW: 69763.5 Da PI: 5.4457 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 154.6 | 7.2e-48 | 8 | 150 | 4 | 142 |
DUF822 4 grkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpleeaeaagssasaspesslq 95
g+k+++kE+E++k+RER+RRai+++++aGLR++Gn++lp+raD+n+Vl+AL+reAGw+ve+DGttyr++ p ++ ++ +s+es+l+
GSVIVT01036911001 8 GKKEREKEKERTKLRERHRRAITSRMLAGLRQYGNFPLPARADMNDVLAALAREAGWTVEADGTTYRQSPPPS----QMATFPVRSVESPLS 95
899****************************************************************776665....788899999999999 PP
DUF822 96 .sslkssalaspvesysaspksssfpspssldsislasa.......asllpvlsv 142
ssl++++ + ++ + ++++ ++++sp+slds+ +++ +s++p++s
GSVIVT01036911001 96 aSSLRNCSAKGSLDCQPSVLRIDESLSPASLDSVVVSERdtksekyTSTSPISSA 150
899********************************99886554444444444444 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 1.4E-48 | 7 | 151 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| SuperFamily | SSF51445 | 5.08E-166 | 171 | 605 | IPR017853 | Glycoside hydrolase superfamily |
| Gene3D | G3DSA:3.20.20.80 | 2.5E-178 | 174 | 605 | IPR013781 | Glycoside hydrolase, catalytic domain |
| Pfam | PF01373 | 4.6E-87 | 196 | 568 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 5.5E-59 | 211 | 225 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 5.5E-59 | 232 | 250 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 5.5E-59 | 254 | 275 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 5.5E-59 | 347 | 369 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 5.5E-59 | 420 | 439 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 5.5E-59 | 454 | 470 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 5.5E-59 | 471 | 482 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 5.5E-59 | 489 | 512 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 5.5E-59 | 527 | 549 | IPR001554 | Glycoside hydrolase, family 14 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000272 | Biological Process | polysaccharide catabolic process | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0048831 | Biological Process | regulation of shoot system development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0016161 | Molecular Function | beta-amylase activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 613 aa Download sequence Send to blast |
MKTVWKNGKK EREKEKERTK LRERHRRAIT SRMLAGLRQY GNFPLPARAD MNDVLAALAR 60 EAGWTVEADG TTYRQSPPPS QMATFPVRSV ESPLSASSLR NCSAKGSLDC QPSVLRIDES 120 LSPASLDSVV VSERDTKSEK YTSTSPISSA ECLEADQLMR DVRSGEHEND FTGTPYVPVY 180 VMLATGVINN FCQLVDPDGI RQELSHMKSL HTDGVVVDCW WGIVEGWSPQ KYEWSGYREL 240 FNIIREFKLK LQVVMAFHEY GGNGSGDVMI SLPQWVLEIG KENQDIFFTD REGRRNTECL 300 SWAIDKERVL KGRTGIEVYF DFMRSFRTEF DDLFAEGIIS AVEIGLGASG ELKYPSFSER 360 MGWAYPGIGE FQCYDKYSQQ NLRKAAKLRG HSFWARGPDN AGQYNSRPHE TGFFCERGDY 420 DSYYGRFFLH WYAQSLIDHA DNVLSLATLA FEETQLIVKV PAVYWWYRTA SHAAELTAGY 480 YNPTNQDGYS PVFEVLKKHS VTMKFVCSGL QITCQENDDA FADPEGLSWQ VLNSAWDRGL 540 TVAGENAVPC YDREGYMRIA EIAKPRNDPD RRHLSFFVYQ QPSPLVERTI WFSELDYFIK 600 CMHGEIAGYL DP* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wdq_A | 1e-129 | 176 | 605 | 11 | 444 | Beta-amylase |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Vvi.4579 | 0.0 | flower| fruit| leaf | ||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00543 | DAP | Transfer from AT5G45300 | Download |
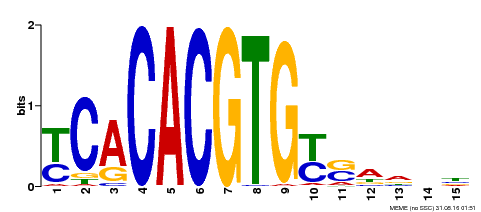
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002270680.1 | 0.0 | PREDICTED: beta-amylase 8 isoform X1 | ||||
| Swissprot | Q9FH80 | 0.0 | BAM8_ARATH; Beta-amylase 8 | ||||
| TrEMBL | F6HJ37 | 0.0 | F6HJ37_VITVI; Beta-amylase | ||||
| STRING | VIT_02s0087g00440.t01 | 0.0 | (Vitis vinifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP9765 | 11 | 12 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45300.1 | 0.0 | beta-amylase 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | GSVIVT01036911001 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



