 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Gh_A03G0607 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Malvales; Malvaceae; Malvoideae; Gossypium
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 419aa MW: 46264.6 Da PI: 6.9524 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 46.6 | 6.3e-15 | 228 | 273 | 5 | 55 |
HHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 5 hnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRR+ri++++++L++l+P+a K +Ka++L +++eY+k+Lq
Gh_A03G0607 228 HNLSERRRRHRIKQKMNTLQKLVPNA-----SKTDKASMLDEVIEYLKQLQ 273
9************************9.....7******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 1.31E-18 | 221 | 292 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 17.212 | 223 | 272 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 2.0E-18 | 228 | 292 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 3.30E-15 | 228 | 277 | No hit | No description |
| Pfam | PF00010 | 1.8E-12 | 228 | 273 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.0E-16 | 229 | 278 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0016607 | Cellular Component | nuclear speck | ||||
| GO:0003690 | Molecular Function | double-stranded DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 419 aa Download sequence Send to blast |
MSHCVVPNCW NLKHQSQEQV HFVEEEENEA NRSSHISNPM SNHEVAELTW QNGQLAFHGL 60 NRHLPTASSK PTWGISGDTL ESIVHQATCL HTQNHNFNLL QHDQNQTPVV VSSIAASSGS 120 SGRSPTAAVV KKRARSSSDV CGEIPRGGVG EEYASACASA NAAFCKENDD STMMTWASHL 180 SPQTMKAKAI DEDYAYQDGS ENQDEEQETR GETDRSQATR RSRASAIHNL SERRRRHRIK 240 QKMNTLQKLV PNASKTDKAS MLDEVIEYLK QLQTQVEMMS MRNMAAQMMM QQTRWQLQMS 300 ALARMGVGMG IGMLDDNNSL APNPSPVTAP THTFLPPPPP TFVTPPPMIP TRFAAAAQAN 360 SDASSHGSIP SPDPYCTFLA QSMNMELYNN KMAALYRPQI NQTTQTSSNP SRSNNVAKD |
| Expression -- Microarray ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | E-value | ||||
| GEO | 73847684 | 0.0 | ||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00571 | DAP | Transfer from AT5G61270 | Download |
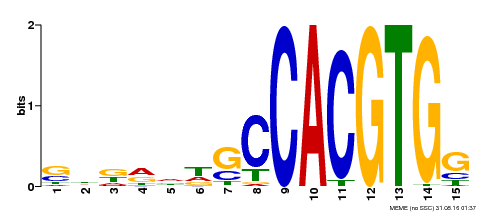
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016665440.1 | 0.0 | PREDICTED: transcription factor PIF7-like | ||||
| TrEMBL | A0A1U8HHA6 | 0.0 | A0A1U8HHA6_GOSHI; transcription factor PIF7-like | ||||
| STRING | Gorai.003G099500.1 | 0.0 | (Gossypium raimondii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3919 | 24 | 51 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00050.1 | 9e-32 | bHLH family protein | ||||



