 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Gh_A03G1332 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Malvales; Malvaceae; Malvoideae; Gossypium
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 255aa MW: 29034 Da PI: 9.3638 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 166.1 | 1.3e-51 | 15 | 139 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpkkvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdkevlsk.kg 97
lppGfrFhPtdeelvv+yL++kv + +l++ ++i+evd++k++PwdLp e+e yfFs+r+ ky++g+r+nrat sgyWkatg dk+++s ++
Gh_A03G1332 15 LPPGFRFHPTDEELVVQYLRRKVLSWPLPA-SIIPEVDVCKADPWDLPGD---LEQERYFFSTREAKYPNGNRSNRATLSGYWKATGIDKQIVSCsGN 108
79****************************.89***************54...46799**********************************997578 PP
NAM 98 elvglkktLvfykgrapkgektdWvmheyrl 128
++vg+kktLvfy+g+ p+g++tdW+mheyrl
Gh_A03G1332 109 QVVGMKKTLVFYRGKPPEGTRTDWIMHEYRL 139
88***************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 1.29E-60 | 10 | 172 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 57.55 | 15 | 172 | IPR003441 | NAC domain |
| Pfam | PF02365 | 1.0E-26 | 16 | 139 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0010089 | Biological Process | xylem development | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 255 aa Download sequence Send to blast |
MMAKLNFVKN GVLRLPPGFR FHPTDEELVV QYLRRKVLSW PLPASIIPEV DVCKADPWDL 60 PGDLEQERYF FSTREAKYPN GNRSNRATLS GYWKATGIDK QIVSCSGNQV VGMKKTLVFY 120 RGKPPEGTRT DWIMHEYRLV TADTCNAPHK KNQTQNHVVA VENWVLCRIF LKKRSGATKR 180 EDDGLQCCNE KGVGKARRKS PVFYEFLSKE RTDLYLAPAS SSSCSSGITQ VSNDNADDHE 240 ESSSCNSFPY FRRKP |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ut4_A | 6e-53 | 13 | 179 | 15 | 171 | NO APICAL MERISTEM PROTEIN |
| 1ut4_B | 6e-53 | 13 | 179 | 15 | 171 | NO APICAL MERISTEM PROTEIN |
| 1ut7_A | 6e-53 | 13 | 179 | 15 | 171 | NO APICAL MERISTEM PROTEIN |
| 1ut7_B | 6e-53 | 13 | 179 | 15 | 171 | NO APICAL MERISTEM PROTEIN |
| 3swm_A | 7e-53 | 13 | 179 | 18 | 174 | NAC domain-containing protein 19 |
| 3swm_B | 7e-53 | 13 | 179 | 18 | 174 | NAC domain-containing protein 19 |
| 3swm_C | 7e-53 | 13 | 179 | 18 | 174 | NAC domain-containing protein 19 |
| 3swm_D | 7e-53 | 13 | 179 | 18 | 174 | NAC domain-containing protein 19 |
| 3swp_A | 7e-53 | 13 | 179 | 18 | 174 | NAC domain-containing protein 19 |
| 3swp_B | 7e-53 | 13 | 179 | 18 | 174 | NAC domain-containing protein 19 |
| 3swp_C | 7e-53 | 13 | 179 | 18 | 174 | NAC domain-containing protein 19 |
| 3swp_D | 7e-53 | 13 | 179 | 18 | 174 | NAC domain-containing protein 19 |
| 4dul_A | 6e-53 | 13 | 179 | 15 | 171 | NAC domain-containing protein 19 |
| 4dul_B | 6e-53 | 13 | 179 | 15 | 171 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Ghi.3574 | 0.0 | boll| ovule | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Up-regulated during xylem vessel element differentiation. {ECO:0000269|PubMed:20388856}. | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in xylem and phloem cells in roots and inflorescence stems (PubMed:20388856). Highly expressed in senescent leaves. Expressed in roots, and abscission and dehiscence tissues, such as axils of bracts and abscission zones in cauline leaves and siliques (PubMed:21673078). {ECO:0000269|PubMed:20388856, ECO:0000269|PubMed:21673078}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional repressor that negatively regulates the expression of genes involved in xylem vessel formation. Represses the transcriptional activation activity of NAC030/VND7, which regulates protoxylem vessel differentiation by promoting immature xylem vessel-specific genes expression (PubMed:20388856). Transcriptional activator that regulates the COLD-REGULATED (COR15A and COR15B) and RESPONSIVE TO DEHYDRATION (LTI78/RD29A and LTI65/RD29B) genes by binding directly to their promoters. Mediates signaling crosstalk between salt stress response and leaf aging process (PubMed:21673078). May play a role in DNA replication of mungbean yellow mosaic virus (PubMed:24442717). {ECO:0000269|PubMed:20388856, ECO:0000269|PubMed:21673078, ECO:0000269|PubMed:24442717}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00507 | DAP | Transfer from AT5G13180 | Download |
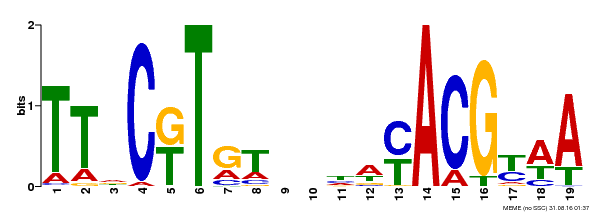
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By abscisic acid (ABA) and salt stress. {ECO:0000269|PubMed:21673078}. | |||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KC110642 | 0.0 | KC110642.1 Gossypium hirsutum NAC protein 7 (NAC7) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016666242.1 | 0.0 | PREDICTED: NAC domain-containing protein 83-like | ||||
| Refseq | XP_017643681.1 | 0.0 | PREDICTED: NAC domain-containing protein 83-like | ||||
| Swissprot | Q9FY93 | 3e-97 | NAC83_ARATH; NAC domain-containing protein 83 | ||||
| TrEMBL | A0A1U8HJJ6 | 0.0 | A0A1U8HJJ6_GOSHI; NAC domain-containing protein 83-like | ||||
| STRING | Gorai.005G195300.1 | 0.0 | (Gossypium raimondii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM1806 | 27 | 86 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G13180.1 | 6e-99 | NAC domain containing protein 83 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



