 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Gh_A05G0554 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Malvales; Malvaceae; Malvoideae; Gossypium
|
||||||||
| Family | EIL | ||||||||
| Protein Properties | Length: 603aa MW: 68073.6 Da PI: 6.2431 | ||||||||
| Description | EIL family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | EIN3 | 431.9 | 6.3e-132 | 40 | 402 | 1 | 337 |
XXXXXXXXXXXXXXXXXXXXXXX..XXXXX.XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX CS
EIN3 1 eelkkrmwkdqmllkrlkerkkqlledkeaatgakksnksneqarrkkmsraQDgiLkYMlkemevcnaqGfvYgiipekgkpvegasdsLraWWkek 98
+el++r wkd+++l+r+ker+k + +a++++ks++ ++qarrkkmsraQDgiLkYMlk mevc+a+GfvYgiipekgkpv+gasd++r+WWkek
Gh_A05G0554 40 DELERRIWKDRIKLRRIKEREKIA---ALQAVEKQKSKQASDQARRKKMSRAQDGILKYMLKLMEVCKARGFVYGIIPEKGKPVSGASDNIRSWWKEK 134
79******************9974...45669****************************************************************** PP
XXXXXXXXXXXXXXXXXXXXXXXXXXX....XX----STTS-HHHHHHHHHHHSSSSSS-TTS--TTT--HHHH---S--HHHHHHT--TT--.---- CS
EIN3 99 vefdrngpaaiskyqaknlilsgesslqtersseshslselqDTtlgSLLsalmqhcdppqrrfplekgvepPWWPtGkelwwgelglskdqgtppyk 196
v+fd+ngpaai+ky+a++l+ s++++ ++ +++ sl++lqD+tlgSLLs+lmqhc+ppqr++plekgv+pPWWPtG+e+ww +lgl++ q+ ppyk
Gh_A05G0554 135 VKFDKNGPAAIAKYEAECLAISESDNI--KKGNSQCSLQDLQDATLGSLLSSLMQHCKPPQRKYPLEKGVPPPWWPTGNEEWWVKLGLQQGQS-PPYK 229
**********************77666..89*************************************************************9.9*** PP
-GGG--HHHHHHHHHHHHHHTGGGHHHHHHTTTTSSSSTTT--SHHHHHHHHHHTTTTT-S--XXXX...XX......XXXXXXXXXXXXXXXXXXXX CS
EIN3 197 kphdlkkawkvsvLtavikhmsptieeirelerqskylqdkmsakesfallsvlnqeekecatvsah...ss......slrkqspkvtlsceqkedve 285
kphdlkk+wkv+vLtavikhmsp+i++ir+++ qsk+lqdkm+akes+++l vl++ee+++++ s + ss r+++++ +s ++++dv+
Gh_A05G0554 230 KPHDLKKMWKVGVLTAVIKHMSPDIAKIRRHVHQSKCLQDKMTAKESAVWLGVLGREEALIQQPSCDkgiSSitekpqGGRDGKKRPAVSIDSDYDVD 327
****************************************************************999665336666665667999999999999**** PP
XXXXXX.XXXXXXXXXX.......................XXXXXXXXXXXXXXXXXXXXX......XXXXXXX. CS
EIN3 286 gkkeskikhvqavktta.......................gfpvvrkrkkkpsesakvsskevsrtcqssqfrgs 337
g ++ + + ++++++++ ++ + rk++++ + + v ++ +++ ++++
Gh_A05G0554 328 GVDDGRGSVSSKDDRRDqpidvepiayinnddshpvqenePVEKQPSRKRPRKGTNHGDRQTVPSHVDKQLVPSH 402
888888888888888778999999999998888888774433344445555555555555555555555555555 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04873 | 1.9E-125 | 40 | 287 | No hit | No description |
| Gene3D | G3DSA:1.10.3180.10 | 4.1E-70 | 163 | 294 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| SuperFamily | SSF116768 | 2.75E-61 | 168 | 290 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042762 | Biological Process | regulation of sulfur metabolic process | ||||
| GO:0071281 | Biological Process | cellular response to iron ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 603 aa Download sequence Send to blast |
MNRVLMVVDL LVDSSDLEVD DIRCDNIAEK DVSDEEIEAD ELERRIWKDR IKLRRIKERE 60 KIAALQAVEK QKSKQASDQA RRKKMSRAQD GILKYMLKLM EVCKARGFVY GIIPEKGKPV 120 SGASDNIRSW WKEKVKFDKN GPAAIAKYEA ECLAISESDN IKKGNSQCSL QDLQDATLGS 180 LLSSLMQHCK PPQRKYPLEK GVPPPWWPTG NEEWWVKLGL QQGQSPPYKK PHDLKKMWKV 240 GVLTAVIKHM SPDIAKIRRH VHQSKCLQDK MTAKESAVWL GVLGREEALI QQPSCDKGIS 300 SITEKPQGGR DGKKRPAVSI DSDYDVDGVD DGRGSVSSKD DRRDQPIDVE PIAYINNDDS 360 HPVQENEPVE KQPSRKRPRK GTNHGDRQTV PSHVDKQLVP SHDEHVNAEL RSPVPDINQT 420 DASFPEYNMP ATQQENDAST MLMHGEKHLI VQSELPAIPC ANEIFTQSMH VDGRPMLYPS 480 VQQTELHHRA AYEFYNPSVE FRHCRDGQQI QVGMNVPQIR PENGIDVSVP AGNDHVITGG 540 ELHHYMKDPF HDEQDRAVHN PFGSPTSDPS FYGGFNSPFE LPFNGTSSLE ELLDDDLIQY 600 FGA |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wij_A | 6e-73 | 167 | 298 | 9 | 140 | ETHYLENE-INSENSITIVE3-like 3 protein |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may be involved in the ethylene response pathway. {ECO:0000269|PubMed:9215635, ECO:0000269|PubMed:9851977}. | |||||
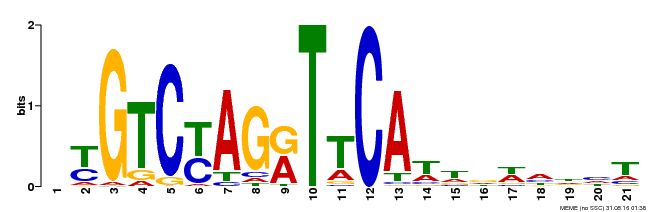
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00233 | DAP | Transfer from AT1G73730 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016750842.1 | 0.0 | PREDICTED: ETHYLENE INSENSITIVE 3-like 3 protein isoform X1 | ||||
| Refseq | XP_016750843.1 | 0.0 | PREDICTED: ETHYLENE INSENSITIVE 3-like 3 protein isoform X1 | ||||
| Refseq | XP_016750844.1 | 0.0 | PREDICTED: ETHYLENE INSENSITIVE 3-like 3 protein isoform X1 | ||||
| Swissprot | O23116 | 0.0 | EIL3_ARATH; ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| TrEMBL | A0A1U8PI16 | 0.0 | A0A1U8PI16_GOSHI; ETHYLENE INSENSITIVE 3-like 3 protein isoform X1 | ||||
| STRING | Gorai.009G071400.1 | 0.0 | (Gossypium raimondii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4658 | 25 | 48 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G73730.1 | 1e-169 | ETHYLENE-INSENSITIVE3-like 3 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



