 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Gh_A05G0559 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Malvales; Malvaceae; Malvoideae; Gossypium
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 424aa MW: 46731 Da PI: 8.1491 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 39.2 | 1.3e-12 | 356 | 401 | 6 | 55 |
HHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 6 nerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
+++Er RR +i +++ +L++l+P+ +k + a++L +Av+YIk+Lq
Gh_A05G0559 356 SIAERVRRTKISERMRKLQDLVPNM----DKQTNTADMLDLAVDYIKDLQ 401
689*********************7....688*****************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50888 | 15.288 | 350 | 400 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 1.31E-16 | 353 | 416 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 4.30E-13 | 354 | 405 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 3.6E-16 | 355 | 420 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 5.6E-14 | 356 | 406 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.5E-9 | 356 | 401 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 424 aa Download sequence Send to blast |
MESDLQHHHH HHLLDYHQPQ HNQKQMNSGL TRYQSAPSSY FSNILDRDFC QEILNRPSSP 60 ETERIIAKFL SSSGDAAGGG DANTENIPPQ NLCTATQSSP VRETPVKIEQ PTQIMTPINN 120 QTGLMQQPNY SSASQNFYQS QPPQYLLNQQ PSASAVDYTT PNPTGMKMGG SNNSNLIRHS 180 SSPAGLFSKI NIENIAGYGV MRGMGDYGSV NSSNREATFR SASRPPTPSG LMTPIAEMGN 240 KSMGPFSSEN AGFGENRPSN YSSGLPVTSW DDSMMISDNM SGIKRLREDD RSLSGLDGAE 300 TKNADGGNHR PPPLLAHHLS LPKSSDMSAI EKFLQYQDSV PCKIRAKRGC ATHPRSIAER 360 VRRTKISERM RKLQDLVPNM DKQTNTADML DLAVDYIKDL QKQVKSLSDS RAKCSCAAQQ 420 QQQQ |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00646 | PBM | Transfer from LOC_Os08g39630 | Download |
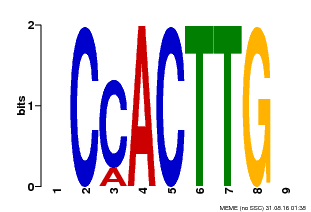
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_017627829.1 | 0.0 | PREDICTED: transcription factor bHLH122 isoform X1 | ||||
| Refseq | XP_017627830.1 | 0.0 | PREDICTED: transcription factor bHLH122 isoform X1 | ||||
| TrEMBL | A0A1U8PI00 | 0.0 | A0A1U8PI00_GOSHI; transcription factor bHLH122-like isoform X1 | ||||
| STRING | Gorai.009G071900.1 | 0.0 | (Gossypium raimondii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM10547 | 26 | 32 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G42280.1 | 6e-63 | bHLH family protein | ||||



