 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Gh_A09G0903 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Malvales; Malvaceae; Malvoideae; Gossypium
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 486aa MW: 54200 Da PI: 7.5228 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 33.2 | 1.1e-10 | 182 | 223 | 4 | 45 |
XCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 4 lkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaL 45
k rr+++NReAAr+sR+RKka++++Le +L++ ++L
Gh_A09G0903 182 AKTLRRLAQNREAARKSRLRKKAYVQQLESSRIKLTQLEQDL 223
6889*************************9777777666655 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 3.3E-8 | 179 | 246 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 9.186 | 181 | 225 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 7.4E-8 | 182 | 224 | IPR004827 | Basic-leucine zipper domain |
| CDD | cd14708 | 1.63E-22 | 183 | 235 | No hit | No description |
| SuperFamily | SSF57959 | 5.79E-7 | 183 | 225 | No hit | No description |
| Gene3D | G3DSA:1.20.5.170 | 2.2E-7 | 184 | 227 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 186 | 201 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF14144 | 3.5E-32 | 264 | 339 | IPR025422 | Transcription factor TGA like domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 486 aa Download sequence Send to blast |
MANHRVGETG LTDSGPSSNH HHHHHNIPYA VLHGMNVPQS FMHQQGPAFD FGELEEAIVL 60 QGVKIRNDEA KPPLFTAGRP AATLEMFPSW PMRFQQTPRG SSKSGGESTD SGSGVNTISS 120 KTENQVEPES PISKKTSSLD QQQQVDMASD ISRTGTSQNQ SAAAKTPQEK RRGSTSEKQL 180 DAKTLRRLAQ NREAARKSRL RKKAYVQQLE SSRIKLTQLE QDLQRARSQG FFLGGCAGTV 240 GNISSGAAIF DMEYSRWLED DQRHMSELRT GLNAHLSDSD LRIIVDSYIS HYDEIFRLKV 300 AAAKADVFHL ITGMWTTPAE RCFLWMGGFR PSDLIKMLIS QLDPLTEQQV MGIYSLQHSS 360 QQAEEALTQG LEQLQQSLID TIAGGPVIDG MQQMAVALGK LANLEGFVRQ ADNLRQQTLH 420 QLSRILTVRQ AARCFLMIGE YYGRLRALSS LWASRPRESM MSEDHSCQTT TELQMVQPSQ 480 NHFSNF |
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: During anther development, accumulates in anther primordia during archesporial cell specification and later present in a horseshoe pattern associated with the lateral and adaxial portion of primordia, prior to the emergence of distinct locules. Expressed throughout sporogenic tissue and surrounding cells layers in adaxial and adaxial locules. Localized to the tapetum and middle layers, gradually fading postmeiosis with degeneration of these cell layers. {ECO:0000269|PubMed:20805327}. | |||||
| Uniprot | TISSUE SPECIFICITY: Mostly expressed in stems, inflorescence apex and flowers, and, to a lower extent, in seedlings, leaves and siliques. {ECO:0000269|PubMed:20805327}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Together with TGA10, basic leucine-zipper transcription factor required for anther development, probably via the activation of SPL expression in anthers and via the regulation of genes with functions in early and middle tapetal development (PubMed:20805327). Required for signaling responses to pathogen-associated molecular patterns (PAMPs) such as flg22 that involves chloroplastic reactive oxygen species (ROS) production and subsequent expression of H(2)O(2)-responsive genes (PubMed:27717447). {ECO:0000269|PubMed:20805327, ECO:0000269|PubMed:27717447}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00131 | DAP | Transfer from AT1G08320 | Download |
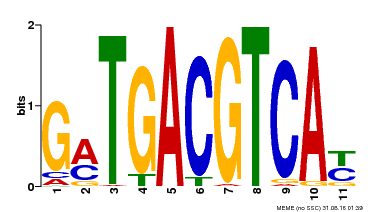
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by flg22 in leaves. {ECO:0000269|PubMed:27717447}. | |||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | JX587228 | 1e-117 | JX587228.1 Gossypium hirsutum clone NBRI_GE21262 microsatellite sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016669014.1 | 0.0 | PREDICTED: transcription factor TGA2-like | ||||
| Swissprot | Q93XM6 | 0.0 | TGA9_ARATH; Transcription factor TGA9 | ||||
| TrEMBL | A0A1U8I048 | 0.0 | A0A1U8I048_GOSHI; transcription factor TGA2-like | ||||
| STRING | Gorai.006G111800.1 | 0.0 | (Gossypium raimondii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM5524 | 24 | 41 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G08320.3 | 0.0 | bZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



