 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Gh_A11G1635 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Malvales; Malvaceae; Malvoideae; Gossypium
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 348aa MW: 40262.1 Da PI: 5.005 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 179.8 | 7.1e-56 | 8 | 137 | 1 | 129 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpk..kvka.eekewyfFskrdkkyatgkrknratksgyWkatgkdkevlsk 95
+ppGfrFhPtdeelv +yL+kkv+++k++l +vi+++d+y++ePwdL++ ++ e++ewyfFs++dkky+tg+r+nrat +g+Wkatg+dk+v++
Gh_A11G1635 8 VPPGFRFHPTDEELVGYYLRKKVASQKIDL-DVIRDIDLYRIEPWDLQErcRIGYeEQSEWYFFSHKDKKYPTGTRTNRATMAGFWKATGRDKAVYD- 103
69****************************.9***************953433332566**************************************. PP
NAM 96 kgelvglkktLvfykgrapkgektdWvmheyrle 129
k++l+g++ktLvfykgrap+g+ktdW+mheyrle
Gh_A11G1635 104 KSKLIGMRKTLVFYKGRAPNGQKTDWIMHEYRLE 137
999*****************************85 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 7.19E-61 | 5 | 157 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 59.711 | 8 | 157 | IPR003441 | NAC domain |
| Pfam | PF02365 | 5.7E-29 | 9 | 136 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0048759 | Biological Process | xylem vessel member cell differentiation | ||||
| GO:1901348 | Biological Process | positive regulation of secondary cell wall biogenesis | ||||
| GO:1990110 | Biological Process | callus formation | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 348 aa Download sequence Send to blast |
MESMESCVPP GFRFHPTDEE LVGYYLRKKV ASQKIDLDVI RDIDLYRIEP WDLQERCRIG 60 YEEQSEWYFF SHKDKKYPTG TRTNRATMAG FWKATGRDKA VYDKSKLIGM RKTLVFYKGR 120 APNGQKTDWI MHEYRLESDE NGPPQEEGWV VCRAFKKKIT GQTKNIEVWD SSYDEPSGIS 180 SVVDPLDYLS RQPQNFLPPS YLCKQETEAE NLNFVHSDQF VQLPQLESPS LPLIKKQTSI 240 SLIPENNNNT NHKEEEDHLL KIMCNNNNNE KVTDWRTLDK FVASQLSHED RYHGEVAVSC 300 FDAADNNINN SDMALLLLQS SGREEANKLN EFLNSSSDCD IGICIFDK |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 3ulx_A | 5e-51 | 5 | 158 | 12 | 169 | Stress-induced transcription factor NAC1 |
| Search in ModeBase | ||||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Up-regulated during xylem vessel element formation. Expressed preferentially in procambial cells adjacent to root meristem. {ECO:0000269|PubMed:16103214, ECO:0000269|PubMed:25148240}. | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in root metaxylem pole and in shoot pre-procambium and procambium (PubMed:18445131). Present in root developing xylems (PubMed:16103214). Specifically expressed in vessels but not in interfascicular fibers in stems (PubMed:25148240). {ECO:0000269|PubMed:16103214, ECO:0000269|PubMed:18445131, ECO:0000269|PubMed:25148240}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the secondary wall NAC binding element (SNBE), 5'-(T/A)NN(C/T)(T/C/G)TNNNNNNNA(A/C)GN(A/C/T)(A/T)-3', in the promoter of target genes (By similarity). Involved in xylem formation by promoting the expression of secondary wall-associated transcription factors and of genes involved in secondary wall biosynthesis and programmed cell death, genes driven by the secondary wall NAC binding element (SNBE). Triggers thickening of secondary walls (PubMed:25148240). {ECO:0000250|UniProtKB:Q9LVA1, ECO:0000269|PubMed:25148240}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00266 | DAP | Transfer from AT2G18060 | Download |
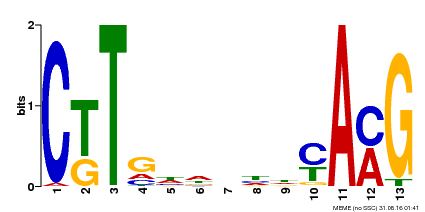
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by chitin (e.g. chitooctaose) (PubMed:17722694). Accumulates in plants exposed to callus induction medium (CIM) (PubMed:17581762). {ECO:0000269|PubMed:17581762, ECO:0000269|PubMed:17722694}. | |||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KC847207 | 0.0 | KC847207.1 Gossypium hirsutum NAC domain protein NAC30 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_017629786.1 | 0.0 | PREDICTED: NAC domain-containing protein 37-like | ||||
| Refseq | XP_017629787.1 | 0.0 | PREDICTED: NAC domain-containing protein 37-like | ||||
| Refseq | XP_017629788.1 | 0.0 | PREDICTED: NAC domain-containing protein 37-like | ||||
| Swissprot | Q9SL41 | 1e-165 | NAC37_ARATH; NAC domain-containing protein 37 | ||||
| TrEMBL | A0A1U8K7H5 | 0.0 | A0A1U8K7H5_GOSHI; NAC domain-containing protein 37-like | ||||
| STRING | Gorai.007G196400.1 | 0.0 | (Gossypium raimondii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3686 | 26 | 61 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G18060.1 | 1e-151 | vascular related NAC-domain protein 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



