 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Gh_A11G1675 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Malvales; Malvaceae; Malvoideae; Gossypium
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 200aa MW: 23021.9 Da PI: 7.7635 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 41 | 4.1e-13 | 65 | 122 | 5 | 62 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklkse 62
++ +r+++NReAArrsR RK++ ++ L v +L eN + +++ ++ + + ++e
Gh_A11G1675 65 RKRKRMASNREAARRSRMRKQKHLDDLMGQVSQLAHENNQILANINITTQIYLNIEAE 122
5789***********************************9999998877777666655 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 1.2E-15 | 61 | 125 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 10.404 | 63 | 110 | IPR004827 | Basic-leucine zipper domain |
| Gene3D | G3DSA:1.20.5.170 | 4.6E-11 | 63 | 140 | No hit | No description |
| Pfam | PF00170 | 4.6E-10 | 65 | 110 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 3.06E-12 | 65 | 117 | No hit | No description |
| CDD | cd14702 | 2.99E-18 | 66 | 116 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 68 | 83 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0046982 | Molecular Function | protein heterodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 200 aa Download sequence Send to blast |
MSPVLCEILL SGLMINSTLR RRTHLVQSFS VVFLYWFYVT GSSSSSATRR SSSSVEEFQQ 60 VLNERKRKRM ASNREAARRS RMRKQKHLDD LMGQVSQLAH ENNQILANIN ITTQIYLNIE 120 AENTVLRAQM AELSTRLQSL NEINDFISSN NGVFGYDHPN NYEADHDHHH HQVNIDTFMN 180 NPWGCFYGNQ PIIASADMIM |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 64 | 85 | RKRKRMASNREAARRSRMRKQK |
| 2 | 77 | 84 | RRSRMRKQ |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Ghi.10464 | 2e-43 | boll| leaf| ovule| stem | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Expressed in the shoot meristem during vegetative to reproductive phase transition. {ECO:0000269|PubMed:22319055}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds to specific DNA sequences in target gene promoters. BZIP2-BZIP63-KIN10 complex binds to the ETFQO promoter to up-regulate its transcription (PubMed:29348240). {ECO:0000250|UniProtKB:O65683, ECO:0000269|PubMed:29348240}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00267 | DAP | Transfer from AT2G18160 | Download |
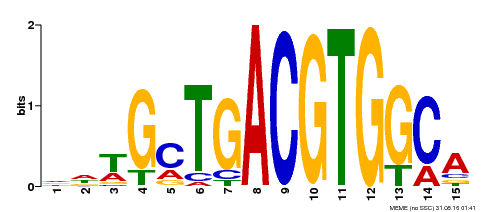
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016682613.1 | 1e-117 | PREDICTED: bZIP transcription factor 53-like | ||||
| Swissprot | Q9SI15 | 1e-42 | BZIP2_ARATH; bZIP transcription factor 2 | ||||
| TrEMBL | A0A0D2TI92 | 1e-125 | A0A0D2TI92_GOSRA; Uncharacterized protein | ||||
| STRING | Gorai.007G200600.1 | 1e-126 | (Gossypium raimondii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM501 | 28 | 154 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G18160.1 | 7e-45 | basic leucine-zipper 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



