 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Gh_A12G1550 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Malvales; Malvaceae; Malvoideae; Gossypium
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 379aa MW: 41985.3 Da PI: 6.7422 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 103.1 | 1.7e-32 | 213 | 268 | 1 | 56 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRla 56
k+r++W+peLH+rF++a++qLGGs++AtPk+i+elmkv+gLt+++vkSHLQkYRl+
Gh_A12G1550 213 KQRRCWSPELHRRFLHALQQLGGSHVATPKQIRELMKVDGLTNDEVKSHLQKYRLH 268
79****************************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 5.9E-29 | 210 | 271 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 14.185 | 210 | 270 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.02E-18 | 211 | 271 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 2.9E-27 | 213 | 268 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 1.5E-7 | 215 | 266 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0055062 | Biological Process | phosphate ion homeostasis | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 379 aa Download sequence Send to blast |
MIEMDYAQKK RRYHEYVEAL EEERRKIQVF QRELPLCLEL VTQAIEAHKK EMSGGSTTTD 60 YLQGQSDCSE QTTSDVPVLE EFIPIKRSSN CSEEDDEQES HKSKETNVPA AVDKRKSDWL 120 RSVQLWNNNQ SPDLPLNEDD DKIRSAVEVK KNGGAFQPFQ KEKTAEMSGQ SAGGKTNGSA 180 TSTCTTESGS RGGEANNANT SKTEEKEGQA SRKQRRCWSP ELHRRFLHAL QQLGGSHVAT 240 PKQIRELMKV DGLTNDEVKS HLQKYRLHTR RPSPTVHNNA NPQAPQFVVV GGIWVPPPDY 300 STMTTTTTST KTATMTPTSG IYTPVAAPLP ALPQPSGTMA QRPQQSDSEE RGSHSEGRVQ 360 SNSPSISSST HTTTDSPAF |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 1e-12 | 213 | 266 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 1e-12 | 213 | 266 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_A | 1e-12 | 213 | 266 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 1e-12 | 213 | 266 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 1e-12 | 213 | 266 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 1e-12 | 213 | 266 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 1e-12 | 213 | 266 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 1e-12 | 213 | 266 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 1e-12 | 213 | 266 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 1e-12 | 213 | 266 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 1e-12 | 213 | 266 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 1e-12 | 213 | 266 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Ghi.501 | 0.0 | boll| ovule | ||||
| Expression -- Microarray ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | E-value | ||||
| GEO | 73863377 | 0.0 | ||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in phosphate homeostasis. Involved in the regulation of the developmental response of lateral roots, acquisition and/or mobilization of phosphate and expression of a subset of genes involved in phosphate sensing and signaling pathway. Is a target of the transcription factor PHR1. {ECO:0000269|PubMed:27016098}. | |||||
| UniProt | Probable transcription factor involved in phosphate signaling in roots. {ECO:0000250|UniProtKB:Q9FX67}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00218 | DAP | Transfer from AT1G68670 | Download |
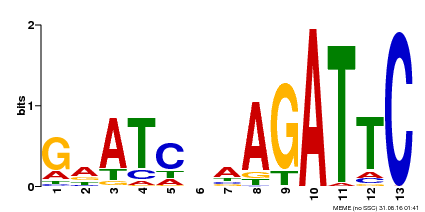
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced under phosphate deprivation conditions. {ECO:0000269|PubMed:27016098}. | |||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HQ526443 | 2e-63 | HQ526443.1 Gossypium herbaceum clone NBRI_D2_1421 simple sequence repeat marker, mRNA sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_017639278.1 | 0.0 | PREDICTED: myb family transcription factor EFM | ||||
| Swissprot | Q8VZS3 | 1e-106 | HHO2_ARATH; Transcription factor HHO2 | ||||
| Swissprot | Q9FPE8 | 1e-106 | HHO3_ARATH; Transcription factor HHO3 | ||||
| TrEMBL | A0A0D2T831 | 0.0 | A0A0D2T831_GOSRA; Uncharacterized protein | ||||
| STRING | Gorai.008G185500.1 | 0.0 | (Gossypium raimondii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3491 | 28 | 63 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G25550.1 | 1e-107 | G2-like family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



