 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Gh_D04G1301 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Malvales; Malvaceae; Malvoideae; Gossypium
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 351aa MW: 39758 Da PI: 8.0512 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 103 | 1.9e-32 | 68 | 121 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
prlrWtp+LH rFv+ave+LGG+++AtPk +l++m++kgL+++hvkSHLQ+YR+
Gh_D04G1301 68 PRLRWTPDLHLRFVHAVERLGGQDRATPKLVLQMMNIKGLSIAHVKSHLQMYRS 121
8****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 12.729 | 64 | 124 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.7E-30 | 64 | 122 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 4.3E-15 | 66 | 122 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 7.2E-23 | 68 | 123 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 5.7E-10 | 69 | 120 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 351 aa Download sequence Send to blast |
MERNSSTECS KTAPSEQSDH QVEAEADSES EENDGESMPR NGGSSSNSTV EENDKKPSVR 60 PYVRSKMPRL RWTPDLHLRF VHAVERLGGQ DRATPKLVLQ MMNIKGLSIA HVKSHLQMYR 120 SKKIDDPRQV ISDHRHLVQS GDGNIFSLNQ LPMLQGYNHH HQGDSSNTFR YRDTSWNGRH 180 FSMRNPYTSR SFTDKQIPVL HGTVTDKIFG SNWTNYNFRM DTSSFNTLLA SWKCHEVLKN 240 EISSSSPNLQ SFLTKPSTQA KVEDETNCSR NSAQERRAMK RTISDSNLDL DLTLRLTQAK 300 EEKRPSSEED DVGLSLSLCG PPSSSKLSRL KGEDDSSREN GRRVSTLDLT I |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4r_A | 5e-17 | 69 | 123 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 5e-17 | 69 | 123 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 5e-17 | 69 | 123 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 5e-17 | 69 | 123 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00297 | DAP | Transfer from AT2G38300 | Download |
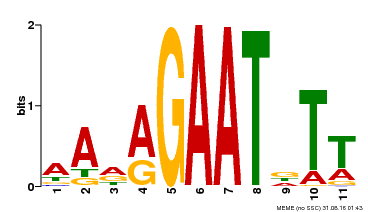
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016680825.1 | 0.0 | PREDICTED: putative two-component response regulator ARR13 isoform X1 | ||||
| TrEMBL | A0A1U8IRY7 | 0.0 | A0A1U8IRY7_GOSHI; putative two-component response regulator ARR13 isoform X1 | ||||
| STRING | Gorai.012G117800.1 | 0.0 | (Gossypium raimondii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM363 | 28 | 184 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G38300.1 | 6e-68 | G2-like family protein | ||||



