 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Gh_D05G1898 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Malvales; Malvaceae; Malvoideae; Gossypium
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 331aa MW: 36791.3 Da PI: 9.2604 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 101.1 | 7.4e-32 | 133 | 186 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
pr+rWt++LH++Fv+av+ LGG+e+AtPk++lelm+vk+Ltl+hvkSHLQ+YR+
Gh_D05G1898 133 PRMRWTTTLHAHFVHAVQLLGGHERATPKSVLELMNVKDLTLAHVKSHLQMYRT 186
9****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.61E-15 | 130 | 187 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 3.7E-28 | 131 | 187 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 2.2E-23 | 133 | 186 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 2.1E-7 | 134 | 185 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0080060 | Biological Process | integument development | ||||
| GO:0005618 | Cellular Component | cell wall | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 331 aa Download sequence Send to blast |
MFLSSNTIMR TAPFPDLSLQ ISPPSVSDCK AKEMAYDVLS GKSIYSDRSS TTDSGSSGSD 60 LSHENGYINP GLGEPTLSLG FEMADLGPPH LQQPRNPHHL HQQHHYQPQI YGRDFKRSAR 120 TMNGGVKRSI RAPRMRWTTT LHAHFVHAVQ LLGGHERATP KSVLELMNVK DLTLAHVKSH 180 LQMYRTVKST DKGSGQGQTD MSLNQRVGIF DLDGRLSSPK ADRNASYSLK LSTPSPQTIP 240 QRTQSDSWPS SMETNNFSVS NRGNGLTFKP NDTKVDGDKA VLHMSDRMKE RLDSSSMSPS 300 DMFLNLEITL GRPSWQMDYA ESSNELTLLK C |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 5e-17 | 134 | 188 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 5e-17 | 134 | 188 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_A | 5e-17 | 134 | 188 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 5e-17 | 134 | 188 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 5e-17 | 134 | 188 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 5e-17 | 134 | 188 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 5e-17 | 134 | 188 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 5e-17 | 134 | 188 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 5e-17 | 134 | 188 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 5e-17 | 134 | 188 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 5e-17 | 134 | 188 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 5e-17 | 134 | 188 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
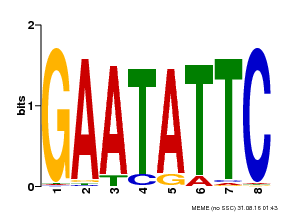
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00107 | PBM | Transfer from AT5G42630 | Download |
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KP271989 | 1e-106 | KP271989.1 UNVERIFIED: Populus trichocarpa isolate Potri.014G037200_BESC-125 mRNA sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_012445063.1 | 0.0 | PREDICTED: probable transcription factor KAN4 | ||||
| TrEMBL | A0A0D2QJW0 | 0.0 | A0A0D2QJW0_GOSRA; Uncharacterized protein | ||||
| STRING | Gorai.009G207200.1 | 0.0 | (Gossypium raimondii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM6572 | 28 | 44 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G42630.1 | 5e-69 | G2-like family protein | ||||



