 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Gh_D05G1946 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Malvales; Malvaceae; Malvoideae; Gossypium
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 458aa MW: 49342.8 Da PI: 10.5422 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 49.6 | 8.8e-16 | 380 | 431 | 5 | 56 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkev 56
+r+rr++kNRe+A rsR+RK+a++ eLe +v++L++eN++L+k+ e+ ++
Gh_D05G1946 380 RRQRRMIKNRESAARSRARKQAYTMELEAEVAKLKEENQELRKKHAEIMEMQ 431
79*****************************************999998875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 6.1E-12 | 376 | 437 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 11.519 | 378 | 429 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 2.6E-13 | 380 | 431 | IPR004827 | Basic-leucine zipper domain |
| Gene3D | G3DSA:1.20.5.170 | 3.6E-15 | 380 | 429 | No hit | No description |
| CDD | cd14707 | 1.07E-27 | 380 | 434 | No hit | No description |
| SuperFamily | SSF57959 | 3.13E-11 | 380 | 429 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 383 | 398 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009738 | Biological Process | abscisic acid-activated signaling pathway | ||||
| GO:0010255 | Biological Process | glucose mediated signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0000976 | Molecular Function | transcription regulatory region sequence-specific DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 458 aa Download sequence Send to blast |
MKKTNKHLAL PRRPKRRPKK QPEAQIILNL KNKILMGTNM NFGSNPPPSG DGGGNKPPGN 60 NLLTRQPSIY SLTFDEFQST MGGIGKDFGS MNMDELLRNI WNTEEIQTMA SSGGVLEGNG 120 GLQRQGSLTL PRTLSQKTVD EVWKDISKEY SLGKDGIGGG GGTNNMPQRQ QTLGEMTLEE 180 FLVRAGVVIE DTQLAGKVNN EGFFGGNNTG FEIGFQQGGK GPNLMGTRIP DGGNQIGIQA 240 SNLHPNVNGV RSNQHQLAQQ HQHQQPIFPK QTGVGYGAQI PLQSGGQLGS PGIRSGMHGI 300 GDQGISNGLI QAGALQGGGM GMVGLGGAVG VATGSPANQV SSDGIGKSSG DTSSVSPVPY 360 VFNGSMRGRK YSAVEKVAER RQRRMIKNRE SAARSRARKQ AYTMELEAEV AKLKEENQEL 420 RKKHAEIMEM QKNQVIEMVD MQQGAKKRCL RRTQTGPW |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 11 | 19 | RRPKRRPKK |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Ghi.9296 | 0.0 | boll| ovule| stem | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in roots, leaves, flowers and siliques but not in seeds. {ECO:0000269|PubMed:11005831, ECO:0000269|PubMed:15361142, ECO:0000269|PubMed:16284313}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Involved in ABA and stress responses and acts as a positive component of glucose signal transduction. Functions as transcriptional activator in the ABA-inducible expression of rd29B. Binds specifically to the ABA-responsive element (ABRE) of the rd29B gene promoter. {ECO:0000269|PubMed:11005831, ECO:0000269|PubMed:15361142, ECO:0000269|PubMed:16284313, ECO:0000269|PubMed:16463099}. | |||||
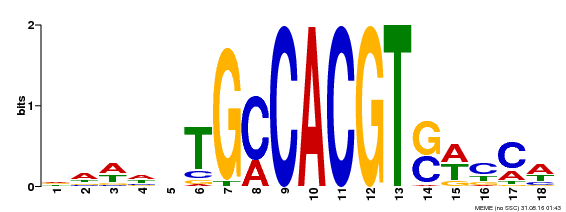
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00186 | DAP | Transfer from AT1G45249 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by drought, salt, abscisic acid (ABA), cold and glucose. {ECO:0000269|PubMed:10636868, ECO:0000269|PubMed:11005831, ECO:0000269|PubMed:15361142, ECO:0000269|PubMed:16284313, ECO:0000269|PubMed:16463099}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016687754.1 | 0.0 | PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 5 | ||||
| Swissprot | Q9M7Q4 | 1e-134 | AI5L5_ARATH; ABSCISIC ACID-INSENSITIVE 5-like protein 5 | ||||
| TrEMBL | A0A1U8JHG4 | 0.0 | A0A1U8JHG4_GOSHI; ABSCISIC ACID-INSENSITIVE 5-like protein 5 | ||||
| STRING | Gorai.009G212600.1 | 0.0 | (Gossypium raimondii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM734 | 27 | 129 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G45249.1 | 1e-107 | abscisic acid responsive elements-binding factor 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



