 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Gh_D08G2447 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Malvales; Malvaceae; Malvoideae; Gossypium
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 351aa MW: 39888.6 Da PI: 7.818 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 174.5 | 3.1e-54 | 16 | 146 | 1 | 129 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLp.k.kvka.eekewyfFskrdkkyatgkrknratksgyWkatgkdkevlsk 95
+ppGfrFhPtdeel+ +yL+kkv+ + ++l +vi+evd++k+ePwdL+ k ++ + ++ewyfFs++dkky+tg+r+nrat++g+Wkatg+dk+++ +
Gh_D08G2447 16 VPPGFRFHPTDEELLYYYLRKKVSYEAIDL-DVIREVDLNKLEPWDLKdKcRIGSgPQNEWYFFSHKDKKYPTGTRTNRATTAGFWKATGRDKAIHLS 112
69****************************.9***************943444443456*************************************** PP
NAM 96 kgelvglkktLvfykgrapkgektdWvmheyrle 129
+++++g++ktLvfy+grap+g+ktdW+mheyrle
Gh_D08G2447 113 NSKKIGMRKTLVFYTGRAPHGQKTDWIMHEYRLE 146
********************************85 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 1.7E-60 | 6 | 165 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 58.396 | 16 | 165 | IPR003441 | NAC domain |
| Pfam | PF02365 | 3.8E-28 | 17 | 145 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003002 | Biological Process | regionalization | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009834 | Biological Process | plant-type secondary cell wall biogenesis | ||||
| GO:0010455 | Biological Process | positive regulation of cell fate commitment | ||||
| GO:0048829 | Biological Process | root cap development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 351 aa Download sequence Send to blast |
MSDINKMMAG NGQLSVPPGF RFHPTDEELL YYYLRKKVSY EAIDLDVIRE VDLNKLEPWD 60 LKDKCRIGSG PQNEWYFFSH KDKKYPTGTR TNRATTAGFW KATGRDKAIH LSNSKKIGMR 120 KTLVFYTGRA PHGQKTDWIM HEYRLEDDDS DVQEDGWVVC RVFKKKNLIR GNFQADFNQE 180 DNNYNHMKNT ASSSAHQMEP RPNNNHLHTL YDYSFDGSMH LPQLFSPESA AVAASSFISP 240 LSLNSMDVEC SQNLLRLTSS SGGGGGGGGV GGGGLMQQDQ MRYNGEWSFL DKLLTNHPMS 300 LDHHHHHHRH PHQSQAKFIP SLQLDHVGSS SQKFPFQYPG CETAEAMKFS K |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ut4_A | 5e-51 | 13 | 167 | 14 | 167 | NO APICAL MERISTEM PROTEIN |
| 1ut4_B | 5e-51 | 13 | 167 | 14 | 167 | NO APICAL MERISTEM PROTEIN |
| 1ut7_A | 5e-51 | 13 | 167 | 14 | 167 | NO APICAL MERISTEM PROTEIN |
| 1ut7_B | 5e-51 | 13 | 167 | 14 | 167 | NO APICAL MERISTEM PROTEIN |
| 4dul_A | 5e-51 | 13 | 167 | 14 | 167 | NAC domain-containing protein 19 |
| 4dul_B | 5e-51 | 13 | 167 | 14 | 167 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: First expressed at early heart stage onward in all root basal daughter cells resulting from horizontal divisions in the COL progenitors and is later maintained in these cells. Present in root stem cell daughters and accumulates in maturing root cap layers. Detectable from very early stages of lateral root development. {ECO:0000269|PubMed:19081078, ECO:0000269|PubMed:20197506}. | |||||
| Uniprot | TISSUE SPECIFICITY: Accumulates in maturing root cap cells, in both COL and LRC cells. {ECO:0000269|PubMed:19081078, ECO:0000269|PubMed:20197506}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription regulator. Together with BRN1 and BRN2, regulates cellular maturation of root cap. Represses stem cell-like divisions in the root cap daughter cells, and thus promotes daughter cell fate. Inhibits expression of its positive regulator FEZ in a feedback loop for controlled switches in cell division plane. Promotes the expression of genes involved in secondary cell walls (SCW) biosynthesis. {ECO:0000269|PubMed:19081078, ECO:0000269|PubMed:20197506}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00250 | DAP | Transfer from AT1G79580 | Download |
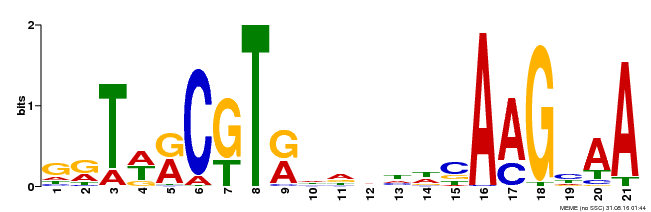
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By FEZ in oriented-divised root cap stem cells. {ECO:0000269|PubMed:19081078}. | |||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | JX617104 | 1e-146 | JX617104.1 Gossypium hirsutum clone NBRI_GE62124 microsatellite sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016714995.1 | 0.0 | PREDICTED: protein SOMBRERO-like | ||||
| Swissprot | Q9MA17 | 1e-122 | SMB_ARATH; Protein SOMBRERO | ||||
| TrEMBL | A0A1U8LKE6 | 0.0 | A0A1U8LKE6_GOSHI; protein SOMBRERO-like | ||||
| STRING | Gorai.004G268500.1 | 0.0 | (Gossypium raimondii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM6653 | 25 | 42 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G79580.3 | 1e-115 | NAC family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



